Gene
KWMTBOMO16460
Pre Gene Modal
BGIBMGA013881
Annotation
hypothetical_protein_RR48_02034_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 3.174
Sequence
CDS
ATGGTCCACTTTATTTTTATAGCATACTTCTGTACGTTGTATCGCGCCATCAACGGCGATAACGAAACTCCCACAGTGGAGAGACGTGAGGATCCGGGAGTACCAGCACAGCATCATCGATATGAGGGTCCGCCCACGCCGTGCCCGGCGCGCGTGGAGTACGCCACTCCAGTGTTCGCGCGGAACTACCAGGGCGTCTGGCGCTACGTCGTGCAGATACCCTACGAGGGGTACTTCACGCAGACCGTCGAGCTCACCAGGTGCATGCAGTCCCGTTGTCATTATTTAGAAGGTGGATGTCTGAGCTCCCCTCGCTGGGTCTCCCTGCTCGTCGCTGAGTTACACTATCCCACAGAGCTTCAGACTCCTCAGCCGGAGTACTCGCAGTACGCGGCCGAGCCCAGCACCGAGAAGCCACCGCACTGCGACGGACACGACGACATGGGCTGCTTCCAGGTGAGGCTCTACTACGACTGGTTCCTGGTTCCCGGCTCCTGCAAGTGTTGGAGGCCCGACTATTTCGCGAGGTATGTCCGGCGCCGTCAGAACAACGAGTTGTAA
Protein
MVHFIFIAYFCTLYRAINGDNETPTVERREDPGVPAQHHRYEGPPTPCPARVEYATPVFARNYQGVWRYVVQIPYEGYFTQTVELTRCMQSRCHYLEGGCLSSPRWVSLLVAELHYPTELQTPQPEYSQYAAEPSTEKPPHCDGHDDMGCFQVRLYYDWFLVPGSCKCWRPDYFARYVRRRQNNEL
Summary
Uniprot
H9JWG7
A0A2H1VGK8
A0A2A4JZK4
A0A2W1BDR1
A0A0N1IQA2
A0A194QG05
+ More
Q9W116 B4QCJ1 B4IH73 B3MF43 Q8I1E1 B4MP86 Q28Z12 B4GIE8 A0A0L0CDK5 A0A1A9ZZH3 A0A0A1WGK0 Q8I199 A0A1A9V6A7 B4LN46 A0A1W4ULT2 B4J4E3 B3NQX8 B4PBU0 A0A1B0GC53 A0A1A9X9M7 A0A1B0BW39 A0A0M4EIG8 A0A0K8UQC5 A0A1I8Q355 B4KTI0 A0A034VK84 D6WPA0 W8BX38 A0A1I8MPN7 A0A1A9WI55 A0A0T6B3V1 Q8MQZ4 A0A3B0JMK4 E2BPK0 A0A195EVF4 E1ZXZ6 N6T571 F4X263 A0A0L7QU95 A0A195DRK1 E9IF50 A0A1I9UVV0 A0A088A1W2 A0A154PSE1 A0A3L8DB25 A0A1Y1LHU7 A0A026WXI1 A0A151X5J0 A0A195CK52 A0A151I6D6 A0A158NAJ4 A0A1J1IRW8 A0A336MFX5 A0A2J7PVT7 A0A0C9R8C7 A0A182GJF6 A0A067R9I1 B0W5G2 A0A182WAB0 A0A1S3D6A2 A0A182G7Q2 A0A182RTP4 A0A1S4FVN4 Q16MW1 A0A182SZL5 A0A182MEK5 W5J7W1 A0A2S2P6Y6 A0A182JCI0 J9JP16 A0A182YNS3 A0A2S2Q7N0 A0A182N7Z8 A0A084W9K0 A0A0A9XI10 A0A182FH89 A7UT83 A0A182VBL3 A0A1S4GN73 A0A182HP64 E0VH58 A0A182XJ98 A0A182LBD7 A0A182PHD9 A0A182U8L7 K7ITR6 A0A0P4VHP9 A0A069DT35 A0A023F2C4 A0A1B0GQM2 Q7Q9P2 A0A0K2T2V2 A0A0M9A0L0 A0A2A3ELT7
Q9W116 B4QCJ1 B4IH73 B3MF43 Q8I1E1 B4MP86 Q28Z12 B4GIE8 A0A0L0CDK5 A0A1A9ZZH3 A0A0A1WGK0 Q8I199 A0A1A9V6A7 B4LN46 A0A1W4ULT2 B4J4E3 B3NQX8 B4PBU0 A0A1B0GC53 A0A1A9X9M7 A0A1B0BW39 A0A0M4EIG8 A0A0K8UQC5 A0A1I8Q355 B4KTI0 A0A034VK84 D6WPA0 W8BX38 A0A1I8MPN7 A0A1A9WI55 A0A0T6B3V1 Q8MQZ4 A0A3B0JMK4 E2BPK0 A0A195EVF4 E1ZXZ6 N6T571 F4X263 A0A0L7QU95 A0A195DRK1 E9IF50 A0A1I9UVV0 A0A088A1W2 A0A154PSE1 A0A3L8DB25 A0A1Y1LHU7 A0A026WXI1 A0A151X5J0 A0A195CK52 A0A151I6D6 A0A158NAJ4 A0A1J1IRW8 A0A336MFX5 A0A2J7PVT7 A0A0C9R8C7 A0A182GJF6 A0A067R9I1 B0W5G2 A0A182WAB0 A0A1S3D6A2 A0A182G7Q2 A0A182RTP4 A0A1S4FVN4 Q16MW1 A0A182SZL5 A0A182MEK5 W5J7W1 A0A2S2P6Y6 A0A182JCI0 J9JP16 A0A182YNS3 A0A2S2Q7N0 A0A182N7Z8 A0A084W9K0 A0A0A9XI10 A0A182FH89 A7UT83 A0A182VBL3 A0A1S4GN73 A0A182HP64 E0VH58 A0A182XJ98 A0A182LBD7 A0A182PHD9 A0A182U8L7 K7ITR6 A0A0P4VHP9 A0A069DT35 A0A023F2C4 A0A1B0GQM2 Q7Q9P2 A0A0K2T2V2 A0A0M9A0L0 A0A2A3ELT7
Pubmed
19121390
28756777
26354079
10731132
12537568
12537572
+ More
12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 22936249 15632085 26108605 25830018 17550304 25348373 18362917 19820115 24495485 25315136 20798317 23537049 21719571 21282665 30249741 28004739 24508170 21347285 26483478 24845553 17510324 20920257 23761445 25244985 24438588 25401762 26823975 12364791 20566863 20966253 20075255 27129103 26334808 25474469
12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 22936249 15632085 26108605 25830018 17550304 25348373 18362917 19820115 24495485 25315136 20798317 23537049 21719571 21282665 30249741 28004739 24508170 21347285 26483478 24845553 17510324 20920257 23761445 25244985 24438588 25401762 26823975 12364791 20566863 20966253 20075255 27129103 26334808 25474469
EMBL
BABH01029399
ODYU01002465
SOQ39990.1
NWSH01000325
PCG77445.1
KZ150111
+ More
PZC73349.1 KQ459670 KPJ20498.1 KQ459054 KPJ03885.1 AE013599 BT001656 AAF47261.1 AAN71411.1 CM000362 CM002911 EDX08607.1 KMY96419.1 CH480838 EDW49249.1 CH902619 EDV37671.2 AY190942 AAO01029.1 CH963849 EDW73925.1 CM000071 EAL25803.2 CH479183 EDW36268.1 JRES01000655 KNC29554.1 GBXI01016754 JAC97537.1 AY190953 AAO01073.1 CH940648 EDW60050.1 CH916367 EDW01625.1 CH954179 EDV57061.1 CM000158 EDW92594.1 CCAG010018996 JXJN01021585 CP012524 ALC40892.1 GDHF01028064 GDHF01023553 JAI24250.1 JAI28761.1 CH933808 EDW08541.1 GAKP01016979 JAC41973.1 KQ971354 EFA07582.2 GAMC01002738 JAC03818.1 LJIG01009930 KRT82096.1 AY122219 AAM52731.1 OUUW01000001 SPP74526.1 GL449633 EFN82404.1 KQ981954 KYN32235.1 GL435158 EFN73936.1 APGK01052264 KB741211 KB632263 ENN72813.1 ERL90947.1 GL888565 EGI59479.1 KQ414735 KOC62190.1 KQ980612 KYN15129.1 GL762758 EFZ20847.1 KU668685 APA05230.1 KQ435127 KZC14821.1 QOIP01000010 RLU17352.1 GEZM01058096 JAV71910.1 KK107077 EZA60533.1 KQ982519 KYQ55549.1 KQ977642 KYN01101.1 KQ976413 KYM90396.1 ADTU01010340 CVRI01000057 CRL02300.1 UFQT01001137 SSX29096.1 NEVH01020943 PNF20448.1 GBYB01012644 JAG82411.1 JXUM01012815 KQ560362 KXJ82824.1 KK852607 KDR20342.1 DS231842 EDS35282.1 JXUM01046774 KQ561490 KXJ78403.1 CH477847 EAT35683.1 AXCM01000364 ADMH02002078 ETN59468.1 GGMR01012598 MBY25217.1 ABLF02019761 GGMS01004418 MBY73621.1 ATLV01021828 KE525324 KFB46894.1 GBHO01023237 GDHC01007946 JAG20367.1 JAQ10683.1 AAAB01008900 EDO64088.1 APCN01002922 DS235161 EEB12714.1 GDKW01002477 JAI54118.1 GBGD01002028 JAC86861.1 GBBI01003563 JAC15149.1 AJVK01017314 EAA09424.3 HACA01002982 CDW20343.1 KQ435776 KOX74900.1 KZ288215 PBC32670.1
PZC73349.1 KQ459670 KPJ20498.1 KQ459054 KPJ03885.1 AE013599 BT001656 AAF47261.1 AAN71411.1 CM000362 CM002911 EDX08607.1 KMY96419.1 CH480838 EDW49249.1 CH902619 EDV37671.2 AY190942 AAO01029.1 CH963849 EDW73925.1 CM000071 EAL25803.2 CH479183 EDW36268.1 JRES01000655 KNC29554.1 GBXI01016754 JAC97537.1 AY190953 AAO01073.1 CH940648 EDW60050.1 CH916367 EDW01625.1 CH954179 EDV57061.1 CM000158 EDW92594.1 CCAG010018996 JXJN01021585 CP012524 ALC40892.1 GDHF01028064 GDHF01023553 JAI24250.1 JAI28761.1 CH933808 EDW08541.1 GAKP01016979 JAC41973.1 KQ971354 EFA07582.2 GAMC01002738 JAC03818.1 LJIG01009930 KRT82096.1 AY122219 AAM52731.1 OUUW01000001 SPP74526.1 GL449633 EFN82404.1 KQ981954 KYN32235.1 GL435158 EFN73936.1 APGK01052264 KB741211 KB632263 ENN72813.1 ERL90947.1 GL888565 EGI59479.1 KQ414735 KOC62190.1 KQ980612 KYN15129.1 GL762758 EFZ20847.1 KU668685 APA05230.1 KQ435127 KZC14821.1 QOIP01000010 RLU17352.1 GEZM01058096 JAV71910.1 KK107077 EZA60533.1 KQ982519 KYQ55549.1 KQ977642 KYN01101.1 KQ976413 KYM90396.1 ADTU01010340 CVRI01000057 CRL02300.1 UFQT01001137 SSX29096.1 NEVH01020943 PNF20448.1 GBYB01012644 JAG82411.1 JXUM01012815 KQ560362 KXJ82824.1 KK852607 KDR20342.1 DS231842 EDS35282.1 JXUM01046774 KQ561490 KXJ78403.1 CH477847 EAT35683.1 AXCM01000364 ADMH02002078 ETN59468.1 GGMR01012598 MBY25217.1 ABLF02019761 GGMS01004418 MBY73621.1 ATLV01021828 KE525324 KFB46894.1 GBHO01023237 GDHC01007946 JAG20367.1 JAQ10683.1 AAAB01008900 EDO64088.1 APCN01002922 DS235161 EEB12714.1 GDKW01002477 JAI54118.1 GBGD01002028 JAC86861.1 GBBI01003563 JAC15149.1 AJVK01017314 EAA09424.3 HACA01002982 CDW20343.1 KQ435776 KOX74900.1 KZ288215 PBC32670.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000000803
UP000000304
+ More
UP000001292 UP000007801 UP000007798 UP000001819 UP000008744 UP000037069 UP000092445 UP000078200 UP000008792 UP000192221 UP000001070 UP000008711 UP000002282 UP000092444 UP000092443 UP000092460 UP000092553 UP000095300 UP000009192 UP000007266 UP000095301 UP000091820 UP000268350 UP000008237 UP000078541 UP000000311 UP000019118 UP000030742 UP000007755 UP000053825 UP000078492 UP000005203 UP000076502 UP000279307 UP000053097 UP000075809 UP000078542 UP000078540 UP000005205 UP000183832 UP000235965 UP000069940 UP000249989 UP000027135 UP000002320 UP000075920 UP000079169 UP000075900 UP000008820 UP000075901 UP000075883 UP000000673 UP000075880 UP000007819 UP000076408 UP000075884 UP000030765 UP000069272 UP000007062 UP000075903 UP000075840 UP000009046 UP000076407 UP000075882 UP000075885 UP000075902 UP000002358 UP000092462 UP000053105 UP000242457
UP000001292 UP000007801 UP000007798 UP000001819 UP000008744 UP000037069 UP000092445 UP000078200 UP000008792 UP000192221 UP000001070 UP000008711 UP000002282 UP000092444 UP000092443 UP000092460 UP000092553 UP000095300 UP000009192 UP000007266 UP000095301 UP000091820 UP000268350 UP000008237 UP000078541 UP000000311 UP000019118 UP000030742 UP000007755 UP000053825 UP000078492 UP000005203 UP000076502 UP000279307 UP000053097 UP000075809 UP000078542 UP000078540 UP000005205 UP000183832 UP000235965 UP000069940 UP000249989 UP000027135 UP000002320 UP000075920 UP000079169 UP000075900 UP000008820 UP000075901 UP000075883 UP000000673 UP000075880 UP000007819 UP000076408 UP000075884 UP000030765 UP000069272 UP000007062 UP000075903 UP000075840 UP000009046 UP000076407 UP000075882 UP000075885 UP000075902 UP000002358 UP000092462 UP000053105 UP000242457
Pfam
PF16077 Spaetzle
SUPFAM
SSF57501
SSF57501
Gene 3D
ProteinModelPortal
H9JWG7
A0A2H1VGK8
A0A2A4JZK4
A0A2W1BDR1
A0A0N1IQA2
A0A194QG05
+ More
Q9W116 B4QCJ1 B4IH73 B3MF43 Q8I1E1 B4MP86 Q28Z12 B4GIE8 A0A0L0CDK5 A0A1A9ZZH3 A0A0A1WGK0 Q8I199 A0A1A9V6A7 B4LN46 A0A1W4ULT2 B4J4E3 B3NQX8 B4PBU0 A0A1B0GC53 A0A1A9X9M7 A0A1B0BW39 A0A0M4EIG8 A0A0K8UQC5 A0A1I8Q355 B4KTI0 A0A034VK84 D6WPA0 W8BX38 A0A1I8MPN7 A0A1A9WI55 A0A0T6B3V1 Q8MQZ4 A0A3B0JMK4 E2BPK0 A0A195EVF4 E1ZXZ6 N6T571 F4X263 A0A0L7QU95 A0A195DRK1 E9IF50 A0A1I9UVV0 A0A088A1W2 A0A154PSE1 A0A3L8DB25 A0A1Y1LHU7 A0A026WXI1 A0A151X5J0 A0A195CK52 A0A151I6D6 A0A158NAJ4 A0A1J1IRW8 A0A336MFX5 A0A2J7PVT7 A0A0C9R8C7 A0A182GJF6 A0A067R9I1 B0W5G2 A0A182WAB0 A0A1S3D6A2 A0A182G7Q2 A0A182RTP4 A0A1S4FVN4 Q16MW1 A0A182SZL5 A0A182MEK5 W5J7W1 A0A2S2P6Y6 A0A182JCI0 J9JP16 A0A182YNS3 A0A2S2Q7N0 A0A182N7Z8 A0A084W9K0 A0A0A9XI10 A0A182FH89 A7UT83 A0A182VBL3 A0A1S4GN73 A0A182HP64 E0VH58 A0A182XJ98 A0A182LBD7 A0A182PHD9 A0A182U8L7 K7ITR6 A0A0P4VHP9 A0A069DT35 A0A023F2C4 A0A1B0GQM2 Q7Q9P2 A0A0K2T2V2 A0A0M9A0L0 A0A2A3ELT7
Q9W116 B4QCJ1 B4IH73 B3MF43 Q8I1E1 B4MP86 Q28Z12 B4GIE8 A0A0L0CDK5 A0A1A9ZZH3 A0A0A1WGK0 Q8I199 A0A1A9V6A7 B4LN46 A0A1W4ULT2 B4J4E3 B3NQX8 B4PBU0 A0A1B0GC53 A0A1A9X9M7 A0A1B0BW39 A0A0M4EIG8 A0A0K8UQC5 A0A1I8Q355 B4KTI0 A0A034VK84 D6WPA0 W8BX38 A0A1I8MPN7 A0A1A9WI55 A0A0T6B3V1 Q8MQZ4 A0A3B0JMK4 E2BPK0 A0A195EVF4 E1ZXZ6 N6T571 F4X263 A0A0L7QU95 A0A195DRK1 E9IF50 A0A1I9UVV0 A0A088A1W2 A0A154PSE1 A0A3L8DB25 A0A1Y1LHU7 A0A026WXI1 A0A151X5J0 A0A195CK52 A0A151I6D6 A0A158NAJ4 A0A1J1IRW8 A0A336MFX5 A0A2J7PVT7 A0A0C9R8C7 A0A182GJF6 A0A067R9I1 B0W5G2 A0A182WAB0 A0A1S3D6A2 A0A182G7Q2 A0A182RTP4 A0A1S4FVN4 Q16MW1 A0A182SZL5 A0A182MEK5 W5J7W1 A0A2S2P6Y6 A0A182JCI0 J9JP16 A0A182YNS3 A0A2S2Q7N0 A0A182N7Z8 A0A084W9K0 A0A0A9XI10 A0A182FH89 A7UT83 A0A182VBL3 A0A1S4GN73 A0A182HP64 E0VH58 A0A182XJ98 A0A182LBD7 A0A182PHD9 A0A182U8L7 K7ITR6 A0A0P4VHP9 A0A069DT35 A0A023F2C4 A0A1B0GQM2 Q7Q9P2 A0A0K2T2V2 A0A0M9A0L0 A0A2A3ELT7
Ontologies
GO
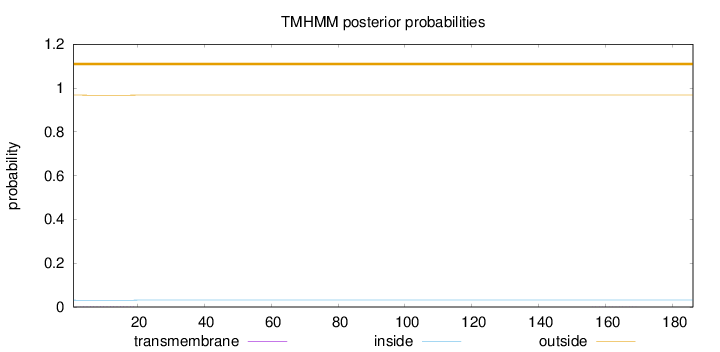
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.864017
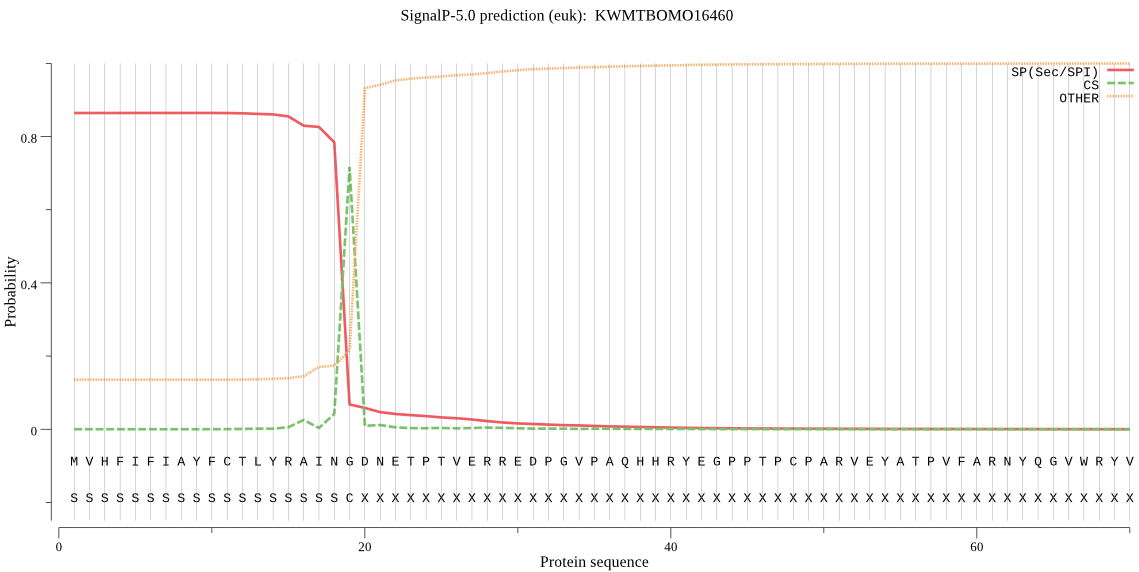
Length:
186
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02998
Exp number, first 60 AAs:
0.02655
Total prob of N-in:
0.03154
outside
1 - 186
Population Genetic Test Statistics
Pi
256.854788
Theta
189.807427
Tajima's D
1.110182
CLR
0
CSRT
0.692215389230538
Interpretation
Uncertain
