Gene
KWMTBOMO16447 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013816
Annotation
PREDICTED:_hydroxylysine_kinase-like?_partial_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 3.163
Sequence
CDS
ATGTGTGGAAGACCTCATAGGTCACATGGTGTTAATTGGTTACCGGAGCCCATAGACATCTACAACAACTTCAACCACTCCGGTCTGGTCAGCAGACAGCACATGTGGATGCTCTCTATGGTCCCAGAGCTGGAGAAATTCAAGTATGTGATCAAGGACTCGGAGAAGCTGGATTTAGCAGAAGAGGTGATAGAAGAATTCAAGTATGCTGTGGTGCCACGCTTGGACGAGCTGGAGAAGGGAGTGATCCATGGTGATATCAATGAGATGAACGTGCTTGTTCTACCGAAGGATGCTGGCGCATCGGATTATCGCATATCAGGAGTGATCGATTTTGGGGACATCCAATATTCGTACTACGTGTTTGAGCTGGCCATCACTATGACGTACATGATGCTGATGTCCGGGGACCCCAGGGTAGGGGGACTGGTGCTCGCCGGCTACACGGTCACCAGGAGGCTGCCTGATGAAGAATACAGACTATTGAAGACCCTGATCTCGGCTCGTCTGGTCCAGAGTTTGATCTTAGGCGCGTACACTCTGGAACAAGACCCGAAAAACACGTATGTCACGTCCACTGAGAAAGCGAATGGATGGGAACTGCTCCGAAAGATCAGGAAAACGAAACCCACAGAGAACGGAGACCCCACGGACTGGAAGGCCATTGCCAATGAGTTTTTGACTAGAAGCTAA
Protein
MCGRPHRSHGVNWLPEPIDIYNNFNHSGLVSRQHMWMLSMVPELEKFKYVIKDSEKLDLAEEVIEEFKYAVVPRLDELEKGVIHGDINEMNVLVLPKDAGASDYRISGVIDFGDIQYSYYVFELAITMTYMMLMSGDPRVGGLVLAGYTVTRRLPDEEYRLLKTLISARLVQSLILGAYTLEQDPKNTYVTSTEKANGWELLRKIRKTKPTENGDPTDWKAIANEFLTRS
Summary
Uniprot
H9JWA2
A0A2H1VFQ9
A0A2A4K060
A0A2W1BEU4
A0A194QK87
A0A0N1PJM1
+ More
A0A212EM13 A0A0L7LT22 Q1DGX8 Q17E72 A0A182GM96 A0A1B0DNP2 A0A182QZM0 A0A1L8DN91 A0A2M4BUF1 A0A2M3ZJ36 A0A2M4CLV5 A0A182FP66 A0A2M4CLV9 A0A182NMK5 A0A2M4BPL2 A0A2M4ARA7 A0A2M4ARI6 A0A2M4ARU1 A0A2M4BPK7 A0A182J1S5 B0W125 A0A182VPQ2 A0A084W7U1 A0A182YEU0 A0A182PTR1 A0A182UNY2 Q7Q7P0 A0A182IDF8 A0A182MPJ6 A0A182U8T6 A0A1Q3F9P8 A0A1Q3F9L4 V5IAL3 W5JAX9 A0A182REB7 A0A182LNM5 A0A182XDQ0 A0A226EUA7 A0A2J7QMS8 A0A0C9R586 A0A034WA15 B4KJQ2 A0A0M4EBC5 A0A0K8UN56 A0A1D2NL90 A0A1Y1M6C7 A0A067RAL0 W8C0S6 A0A1B6CDI4 A0A139WEZ1 A0A0J9R4A9 Q9VJ68 Q8SY12 E9GVP7 A0A1W4XBT9 B4MZN0 B4I5J0 A0A164ZW81 A0A0L0BUH0 A0A1J1J5K1 A0A0P5JHE2 A0A0P5FVN6 A0A1W4VLD4 B4LSK0 B4JBY5 A0A0P5W0E8 U5EXQ9 B4Q8W4 A0A0P5XBB6 A0A0P5VCD4 A0A0P4YQI0 A0A0P5M3W7 A0A0P6HBC9 A0A1B0AUR9 B3NLB4 A0A0P5ST21 A0A0P5ZMF8 A0A0P5TPF0 A0A0P6BY61 A0A0P5J4X4 A0A1A9YBL3 A0A151IQN5 A0A0P5J3F4 A0A0P5GDP9 A0A0P5W2K6 A0A0P4Z3X6 A0A0N8AQ38 A0A1D2NHU1 A0A3B0JFF1 B4P9W3 B3MM40 A0A1A9UL53 T1PHJ4 A0A1A9ZD01
A0A212EM13 A0A0L7LT22 Q1DGX8 Q17E72 A0A182GM96 A0A1B0DNP2 A0A182QZM0 A0A1L8DN91 A0A2M4BUF1 A0A2M3ZJ36 A0A2M4CLV5 A0A182FP66 A0A2M4CLV9 A0A182NMK5 A0A2M4BPL2 A0A2M4ARA7 A0A2M4ARI6 A0A2M4ARU1 A0A2M4BPK7 A0A182J1S5 B0W125 A0A182VPQ2 A0A084W7U1 A0A182YEU0 A0A182PTR1 A0A182UNY2 Q7Q7P0 A0A182IDF8 A0A182MPJ6 A0A182U8T6 A0A1Q3F9P8 A0A1Q3F9L4 V5IAL3 W5JAX9 A0A182REB7 A0A182LNM5 A0A182XDQ0 A0A226EUA7 A0A2J7QMS8 A0A0C9R586 A0A034WA15 B4KJQ2 A0A0M4EBC5 A0A0K8UN56 A0A1D2NL90 A0A1Y1M6C7 A0A067RAL0 W8C0S6 A0A1B6CDI4 A0A139WEZ1 A0A0J9R4A9 Q9VJ68 Q8SY12 E9GVP7 A0A1W4XBT9 B4MZN0 B4I5J0 A0A164ZW81 A0A0L0BUH0 A0A1J1J5K1 A0A0P5JHE2 A0A0P5FVN6 A0A1W4VLD4 B4LSK0 B4JBY5 A0A0P5W0E8 U5EXQ9 B4Q8W4 A0A0P5XBB6 A0A0P5VCD4 A0A0P4YQI0 A0A0P5M3W7 A0A0P6HBC9 A0A1B0AUR9 B3NLB4 A0A0P5ST21 A0A0P5ZMF8 A0A0P5TPF0 A0A0P6BY61 A0A0P5J4X4 A0A1A9YBL3 A0A151IQN5 A0A0P5J3F4 A0A0P5GDP9 A0A0P5W2K6 A0A0P4Z3X6 A0A0N8AQ38 A0A1D2NHU1 A0A3B0JFF1 B4P9W3 B3MM40 A0A1A9UL53 T1PHJ4 A0A1A9ZD01
Pubmed
19121390
28756777
26354079
22118469
26227816
17510324
+ More
26483478 24438588 25244985 12364791 14747013 17210077 20920257 23761445 20966253 25348373 17994087 18057021 27289101 28004739 24845553 24495485 18362917 19820115 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21292972 26108605 17550304 25315136
26483478 24438588 25244985 12364791 14747013 17210077 20920257 23761445 20966253 25348373 17994087 18057021 27289101 28004739 24845553 24495485 18362917 19820115 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21292972 26108605 17550304 25315136
EMBL
BABH01029383
BABH01029384
BABH01029385
ODYU01002327
SOQ39678.1
NWSH01000325
+ More
PCG77439.1 KZ150111 PZC73338.1 KQ459054 KPJ03871.1 KQ459670 KPJ20509.1 AGBW02013941 OWR42534.1 JTDY01000148 KOB78592.1 CH900075 EAT32408.1 CH477285 EAT44747.1 JXUM01074043 JXUM01074044 JXUM01074045 JXUM01074046 JXUM01074047 KQ562809 KXJ75081.1 AJVK01007638 AJVK01007639 AJVK01007640 AXCN02001594 GFDF01006162 JAV07922.1 GGFJ01007538 MBW56679.1 GGFM01007771 MBW28522.1 GGFL01002138 MBW66316.1 GGFL01002139 MBW66317.1 GGFJ01005876 MBW55017.1 GGFK01010006 MBW43327.1 GGFK01010086 MBW43407.1 GGFK01010121 MBW43442.1 GGFJ01005875 MBW55016.1 DS231819 EDS42842.1 ATLV01021298 KE525316 KFB46285.1 AAAB01008952 EAA10548.4 APCN01005972 AXCM01000109 GFDL01010806 JAV24239.1 GFDL01010809 JAV24236.1 GALX01000985 JAB67481.1 ADMH02001905 ETN60563.1 LNIX01000002 OXA61109.1 NEVH01013202 PNF29895.1 GBYB01002006 JAG71773.1 GAKP01008339 GAKP01008338 GAKP01008337 JAC50613.1 CH933807 EDW11497.1 KRG02723.1 CP012523 ALC38710.1 GDHF01024197 GDHF01012677 GDHF01001438 JAI28117.1 JAI39637.1 JAI50876.1 LJIJ01000012 ODN06024.1 GEZM01039696 JAV81284.1 KK852582 KDR20869.1 GAMC01003858 GAMC01003857 JAC02699.1 GEDC01025873 JAS11425.1 KQ971354 KYB26429.1 CM002910 KMY90856.1 AE014134 BT133492 AAF53686.2 AAF53688.2 AFI71922.1 API64994.1 AY075459 AAL68272.1 GL732568 EFX76488.1 CH963920 EDW77815.1 KRF98748.1 CH480822 EDW55646.1 LRGB01000687 KZS16858.1 JRES01001454 KNC22854.1 CVRI01000067 CRL06750.1 GDIQ01205217 GDIQ01205214 JAK46508.1 GDIQ01249704 GDIQ01249702 JAK02021.1 CH940649 EDW64822.1 KRF81885.1 KRF81886.1 CH916368 EDW04088.1 GDIP01093550 JAM10165.1 GANO01000825 JAB59046.1 CM000361 EDX05396.1 KMY90853.1 GDIP01074610 JAM29105.1 GDIP01101905 JAM01810.1 GDIP01228383 JAI95018.1 GDIQ01161281 JAK90444.1 GDIQ01033983 JAN60754.1 JXJN01003720 CH954179 EDV54764.1 GDIP01135806 JAL67908.1 GDIP01055122 JAM48593.1 GDIP01126653 JAL77061.1 GDIP01007731 JAM95984.1 GDIQ01205216 JAK46509.1 KQ976768 KYN08467.1 GDIQ01205215 JAK46510.1 GDIQ01249703 JAK02022.1 GDIP01091944 JAM11771.1 GDIP01219797 JAJ03605.1 GDIQ01253955 JAJ97769.1 LJIJ01000035 ODN04830.1 OUUW01000004 SPP79413.1 CM000158 EDW90304.1 KRJ99113.1 CH902620 EDV30855.1 KPU73084.1 KPU73085.1 KA647590 AFP62219.1
PCG77439.1 KZ150111 PZC73338.1 KQ459054 KPJ03871.1 KQ459670 KPJ20509.1 AGBW02013941 OWR42534.1 JTDY01000148 KOB78592.1 CH900075 EAT32408.1 CH477285 EAT44747.1 JXUM01074043 JXUM01074044 JXUM01074045 JXUM01074046 JXUM01074047 KQ562809 KXJ75081.1 AJVK01007638 AJVK01007639 AJVK01007640 AXCN02001594 GFDF01006162 JAV07922.1 GGFJ01007538 MBW56679.1 GGFM01007771 MBW28522.1 GGFL01002138 MBW66316.1 GGFL01002139 MBW66317.1 GGFJ01005876 MBW55017.1 GGFK01010006 MBW43327.1 GGFK01010086 MBW43407.1 GGFK01010121 MBW43442.1 GGFJ01005875 MBW55016.1 DS231819 EDS42842.1 ATLV01021298 KE525316 KFB46285.1 AAAB01008952 EAA10548.4 APCN01005972 AXCM01000109 GFDL01010806 JAV24239.1 GFDL01010809 JAV24236.1 GALX01000985 JAB67481.1 ADMH02001905 ETN60563.1 LNIX01000002 OXA61109.1 NEVH01013202 PNF29895.1 GBYB01002006 JAG71773.1 GAKP01008339 GAKP01008338 GAKP01008337 JAC50613.1 CH933807 EDW11497.1 KRG02723.1 CP012523 ALC38710.1 GDHF01024197 GDHF01012677 GDHF01001438 JAI28117.1 JAI39637.1 JAI50876.1 LJIJ01000012 ODN06024.1 GEZM01039696 JAV81284.1 KK852582 KDR20869.1 GAMC01003858 GAMC01003857 JAC02699.1 GEDC01025873 JAS11425.1 KQ971354 KYB26429.1 CM002910 KMY90856.1 AE014134 BT133492 AAF53686.2 AAF53688.2 AFI71922.1 API64994.1 AY075459 AAL68272.1 GL732568 EFX76488.1 CH963920 EDW77815.1 KRF98748.1 CH480822 EDW55646.1 LRGB01000687 KZS16858.1 JRES01001454 KNC22854.1 CVRI01000067 CRL06750.1 GDIQ01205217 GDIQ01205214 JAK46508.1 GDIQ01249704 GDIQ01249702 JAK02021.1 CH940649 EDW64822.1 KRF81885.1 KRF81886.1 CH916368 EDW04088.1 GDIP01093550 JAM10165.1 GANO01000825 JAB59046.1 CM000361 EDX05396.1 KMY90853.1 GDIP01074610 JAM29105.1 GDIP01101905 JAM01810.1 GDIP01228383 JAI95018.1 GDIQ01161281 JAK90444.1 GDIQ01033983 JAN60754.1 JXJN01003720 CH954179 EDV54764.1 GDIP01135806 JAL67908.1 GDIP01055122 JAM48593.1 GDIP01126653 JAL77061.1 GDIP01007731 JAM95984.1 GDIQ01205216 JAK46509.1 KQ976768 KYN08467.1 GDIQ01205215 JAK46510.1 GDIQ01249703 JAK02022.1 GDIP01091944 JAM11771.1 GDIP01219797 JAJ03605.1 GDIQ01253955 JAJ97769.1 LJIJ01000035 ODN04830.1 OUUW01000004 SPP79413.1 CM000158 EDW90304.1 KRJ99113.1 CH902620 EDV30855.1 KPU73084.1 KPU73085.1 KA647590 AFP62219.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000008820 UP000069940 UP000249989 UP000092462 UP000075886 UP000069272 UP000075884 UP000075880 UP000002320 UP000075920 UP000030765 UP000076408 UP000075885 UP000075903 UP000007062 UP000075840 UP000075883 UP000075902 UP000000673 UP000075900 UP000075882 UP000076407 UP000198287 UP000235965 UP000009192 UP000092553 UP000094527 UP000027135 UP000007266 UP000000803 UP000000305 UP000192223 UP000007798 UP000001292 UP000076858 UP000037069 UP000183832 UP000192221 UP000008792 UP000001070 UP000000304 UP000092460 UP000008711 UP000092443 UP000078542 UP000268350 UP000002282 UP000007801 UP000078200 UP000095301 UP000092445
UP000008820 UP000069940 UP000249989 UP000092462 UP000075886 UP000069272 UP000075884 UP000075880 UP000002320 UP000075920 UP000030765 UP000076408 UP000075885 UP000075903 UP000007062 UP000075840 UP000075883 UP000075902 UP000000673 UP000075900 UP000075882 UP000076407 UP000198287 UP000235965 UP000009192 UP000092553 UP000094527 UP000027135 UP000007266 UP000000803 UP000000305 UP000192223 UP000007798 UP000001292 UP000076858 UP000037069 UP000183832 UP000192221 UP000008792 UP000001070 UP000000304 UP000092460 UP000008711 UP000092443 UP000078542 UP000268350 UP000002282 UP000007801 UP000078200 UP000095301 UP000092445
Pfam
PF01636 APH
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9JWA2
A0A2H1VFQ9
A0A2A4K060
A0A2W1BEU4
A0A194QK87
A0A0N1PJM1
+ More
A0A212EM13 A0A0L7LT22 Q1DGX8 Q17E72 A0A182GM96 A0A1B0DNP2 A0A182QZM0 A0A1L8DN91 A0A2M4BUF1 A0A2M3ZJ36 A0A2M4CLV5 A0A182FP66 A0A2M4CLV9 A0A182NMK5 A0A2M4BPL2 A0A2M4ARA7 A0A2M4ARI6 A0A2M4ARU1 A0A2M4BPK7 A0A182J1S5 B0W125 A0A182VPQ2 A0A084W7U1 A0A182YEU0 A0A182PTR1 A0A182UNY2 Q7Q7P0 A0A182IDF8 A0A182MPJ6 A0A182U8T6 A0A1Q3F9P8 A0A1Q3F9L4 V5IAL3 W5JAX9 A0A182REB7 A0A182LNM5 A0A182XDQ0 A0A226EUA7 A0A2J7QMS8 A0A0C9R586 A0A034WA15 B4KJQ2 A0A0M4EBC5 A0A0K8UN56 A0A1D2NL90 A0A1Y1M6C7 A0A067RAL0 W8C0S6 A0A1B6CDI4 A0A139WEZ1 A0A0J9R4A9 Q9VJ68 Q8SY12 E9GVP7 A0A1W4XBT9 B4MZN0 B4I5J0 A0A164ZW81 A0A0L0BUH0 A0A1J1J5K1 A0A0P5JHE2 A0A0P5FVN6 A0A1W4VLD4 B4LSK0 B4JBY5 A0A0P5W0E8 U5EXQ9 B4Q8W4 A0A0P5XBB6 A0A0P5VCD4 A0A0P4YQI0 A0A0P5M3W7 A0A0P6HBC9 A0A1B0AUR9 B3NLB4 A0A0P5ST21 A0A0P5ZMF8 A0A0P5TPF0 A0A0P6BY61 A0A0P5J4X4 A0A1A9YBL3 A0A151IQN5 A0A0P5J3F4 A0A0P5GDP9 A0A0P5W2K6 A0A0P4Z3X6 A0A0N8AQ38 A0A1D2NHU1 A0A3B0JFF1 B4P9W3 B3MM40 A0A1A9UL53 T1PHJ4 A0A1A9ZD01
A0A212EM13 A0A0L7LT22 Q1DGX8 Q17E72 A0A182GM96 A0A1B0DNP2 A0A182QZM0 A0A1L8DN91 A0A2M4BUF1 A0A2M3ZJ36 A0A2M4CLV5 A0A182FP66 A0A2M4CLV9 A0A182NMK5 A0A2M4BPL2 A0A2M4ARA7 A0A2M4ARI6 A0A2M4ARU1 A0A2M4BPK7 A0A182J1S5 B0W125 A0A182VPQ2 A0A084W7U1 A0A182YEU0 A0A182PTR1 A0A182UNY2 Q7Q7P0 A0A182IDF8 A0A182MPJ6 A0A182U8T6 A0A1Q3F9P8 A0A1Q3F9L4 V5IAL3 W5JAX9 A0A182REB7 A0A182LNM5 A0A182XDQ0 A0A226EUA7 A0A2J7QMS8 A0A0C9R586 A0A034WA15 B4KJQ2 A0A0M4EBC5 A0A0K8UN56 A0A1D2NL90 A0A1Y1M6C7 A0A067RAL0 W8C0S6 A0A1B6CDI4 A0A139WEZ1 A0A0J9R4A9 Q9VJ68 Q8SY12 E9GVP7 A0A1W4XBT9 B4MZN0 B4I5J0 A0A164ZW81 A0A0L0BUH0 A0A1J1J5K1 A0A0P5JHE2 A0A0P5FVN6 A0A1W4VLD4 B4LSK0 B4JBY5 A0A0P5W0E8 U5EXQ9 B4Q8W4 A0A0P5XBB6 A0A0P5VCD4 A0A0P4YQI0 A0A0P5M3W7 A0A0P6HBC9 A0A1B0AUR9 B3NLB4 A0A0P5ST21 A0A0P5ZMF8 A0A0P5TPF0 A0A0P6BY61 A0A0P5J4X4 A0A1A9YBL3 A0A151IQN5 A0A0P5J3F4 A0A0P5GDP9 A0A0P5W2K6 A0A0P4Z3X6 A0A0N8AQ38 A0A1D2NHU1 A0A3B0JFF1 B4P9W3 B3MM40 A0A1A9UL53 T1PHJ4 A0A1A9ZD01
PDB
6EF6
E-value=0.00428878,
Score=92
Ontologies
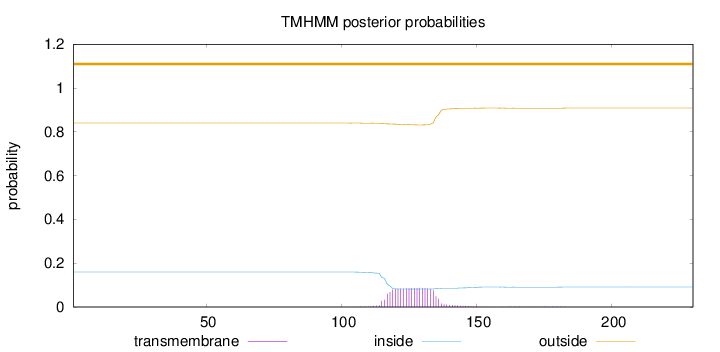
Topology
Length:
230
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.79596
Exp number, first 60 AAs:
0.00056
Total prob of N-in:
0.15995
outside
1 - 230
Population Genetic Test Statistics
Pi
179.758576
Theta
134.887991
Tajima's D
1.167548
CLR
2.25111
CSRT
0.707214639268037
Interpretation
Uncertain
