Gene
KWMTBOMO16444
Pre Gene Modal
BGIBMGA013820
Annotation
PREDICTED:_ATP-sensitive_inward_rectifier_potassium_channel_11-like_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 2.508
Sequence
CDS
ATGGAGGCCAAAGATATACTCAATAATTCAATGGAAAAAGACTCTTTGAAAGCTCAAATGGAATACGACGAAATAGATAAACTTTACGATGAGTGCCTAGAATATTTGAAAGAAGCCAAAATAGACGTTCAAGAAGAAGACCAAGCTCAACACGAAGACCAAGTTGAACAAGAAGAATACATAGAAGACCACGAAAGAATAAAAGAACATAGTGCTAATTCTAGAAACTCAACAGAAACACCAAAAACATCTATCAAGTTGGAAAACGAACTGAATTCGACAGGCAGTCTGGTTTCTGTAATTTCCGAACCAAAATCTGAGAGACTAAGGAAGCTATCGCAGCAGCTGCCTAAAATTATAATCACTCAGAGTAATACAAGTTTGAACTTTAAAAGGTTAGAGGAAGCAGTCGAAATTCCAGTTAAAATAGAGAAGAAAAAGATAAATAGGAATCCTCCGGCTATCCGCAATAAGACTGTTTACGGAGATTTCCCCAAAAGACTCGTCTCCAAGAACGGGATGGAGAACATCGCTCTCATCAAAAACGTACCTTTTGAAAAAATCAGACTCCTCAACGACACCTTCCATACTTTGATCAATCTTCGATGGCGTTGGATCGTCATCGGTACAATGGTGGTCCATTTCGCCATTTGGCTGGTCTTCGCGGTATTCTGGTATATATCAGCATTGGCCCACTACGACTTCGAGCCAGATCCACCGAAGAAGACTTGCGTGACGGGTGGCAAAGATTTCGTGACGATATTCTTGGCATCTGTTGAGGCTCAGACAACGATAGGCTTTGGAGATCGCGCCATAGACGAAGAATGTCCTGAATCTATCTTCCTATTCATCATGCAGTTGATCCTCGGCACCGGGCTATCCGGGGTACTGACATGTGTGGTATACGCAAAACTAACGCGCCCAGAGAAACTGTCTCGCGATGGAGTTGGTTTCAGTAAGAAGGCTGTGATCAGCATGCGCGACGGTCAGCTATGTCTGATGATGAGGGCCTGGGACCTCAACTACGACCACATCATCTGCTCAGAATTCTCAGCACATTTCATAACAACCCACAGGACCAAAGAAGGTGAGGTGTTGCGGTATTACGCCAGGCCTATGGCGCTCCAGCAGCGACAGTTCCCTCTGTGGCCAGTCACTATAGTCCACATTATTGATGCCGGCAGCCCTCTATATGAGTATACTCCAGAAGCTCTGGCGAATAGCAGCTATGAGATTACTTTGAGCCTGAAGGGGGCATCAGCATCAATGGGAACATTTACACAGAGTCAAACGTCTTATCTGCCACGGGAAGTCGTGTGGGGACATAGATTCAAACCAGTTGTCAGATATGACGGCAGAAAACAGAAGTACGTGATTGAAGAGAAGAAACTGGACGCGACAGAGCCTGTTGATACTCCCTACTGTAGTGCCAAGGAATTACACAGTACATCGAAGGATAGCTGCTCGACCCCACGCCCGTGTACTCCAAGCTCCTTCTGCGAAAAGATCTCTGCATTCCAACTGGAAAGATCATCGTCTTTCAGAAAGAAGACCACGGCATCATGA
Protein
MEAKDILNNSMEKDSLKAQMEYDEIDKLYDECLEYLKEAKIDVQEEDQAQHEDQVEQEEYIEDHERIKEHSANSRNSTETPKTSIKLENELNSTGSLVSVISEPKSERLRKLSQQLPKIIITQSNTSLNFKRLEEAVEIPVKIEKKKINRNPPAIRNKTVYGDFPKRLVSKNGMENIALIKNVPFEKIRLLNDTFHTLINLRWRWIVIGTMVVHFAIWLVFAVFWYISALAHYDFEPDPPKKTCVTGGKDFVTIFLASVEAQTTIGFGDRAIDEECPESIFLFIMQLILGTGLSGVLTCVVYAKLTRPEKLSRDGVGFSKKAVISMRDGQLCLMMRAWDLNYDHIICSEFSAHFITTHRTKEGEVLRYYARPMALQQRQFPLWPVTIVHIIDAGSPLYEYTPEALANSSYEITLSLKGASASMGTFTQSQTSYLPREVVWGHRFKPVVRYDGRKQKYVIEEKKLDATEPVDTPYCSAKELHSTSKDSCSTPRPCTPSSFCEKISAFQLERSSSFRKKTTAS
Summary
Similarity
Belongs to the inward rectifier-type potassium channel (TC 1.A.2.1) family.
Uniprot
EMBL
Proteomes
PRIDE
Interpro
SUPFAM
SSF81296
SSF81296
Gene 3D
ProteinModelPortal
PDB
3SPI
E-value=6.9816e-50,
Score=499
Ontologies
KEGG
GO
PANTHER
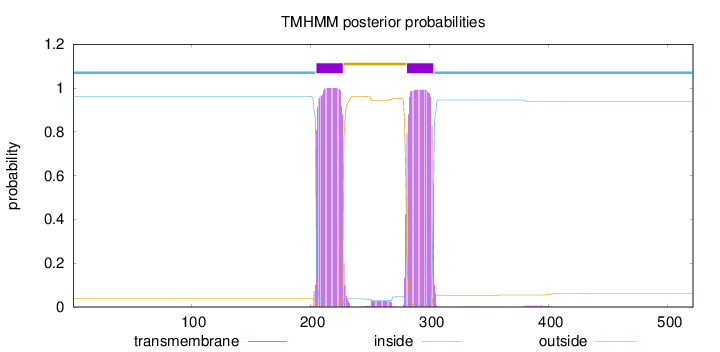
Topology
Subcellular location
Membrane
Length:
521
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
46.09138
Exp number, first 60 AAs:
0
Total prob of N-in:
0.96232
inside
1 - 204
TMhelix
205 - 227
outside
228 - 280
TMhelix
281 - 303
inside
304 - 521
Population Genetic Test Statistics
Pi
323.13722
Theta
169.699448
Tajima's D
3.109769
CLR
0.095996
CSRT
0.984000799960002
Interpretation
Uncertain
