Gene
KWMTBOMO16440
Pre Gene Modal
BGIBMGA013823
Annotation
PREDICTED:_ELAV-like_protein_2_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 0.911
Sequence
CDS
ATGTCGAAGGGCGACAGCGAGCAGCAGAACGGCTCCGGCGAGGAGTCCAAGACCAATCTGATCATCAACTACCTGCCCCAGAGCATGACCCAGGAAGAGATCCGCAGCCTCTTCTCGAGCATCGGTGAAGTGGAATCTTGCAAGTTGATCCGGAACAAGGGCGCGGCCTTCCCCGACGCGCTCAATCACGCTCTGCACGGCGGCGGACAGAGCCTCGGCTACGCCTTTGTCAACTATCACCGGCCCGAGGATGCGGAGAAGGCTATTGCGACCCTCAACGGCCTGCGTCTCCAGAACAAAACAATCAAAGTGTCGTACGCGCGCCCCAGCAGCGAGGCCATCAAAGGTGCCAATTTATACGTATCCGGTCTTCCCAAGACCATGACTCAGAGCGAGCTCGAGCGCCTGTTCAGTCCGTACGGGCGCATCATCACGTCGCGTATACTATGCGAGAACTCGGGCGGGCGGCCCTTCACCGGCGGCGAACAAGGTTTGTCCAAGGGCGTCGGCTTCATCCGCTTCGACCAGCGCGTGGAAGCGGAGCGAGCCATACAAGAGTTGAACGGCACCATACCGAAAGGAGCCAGCGAGCCTATAACAGTGAAGTTCGCGAATAACCCGAGCAACAACGGGAAGGCGCTAGCTCCGTTGGCGGCGTACCTACCAGCCGCGCTGCGGTTCCCCGCGCCGCTGGGCCGGTTCAGTTCAGGCAAGTCGCTTCTCGCTATAAACAAAGGATTACAGCGCTACAGCCCTCTCGCCGGCGAGTTACTCGGAGGGGTCCTGCCCGGCGCCGTGGGCTCGGAGTGGTGCATCTTCGTCTACAACTTAGCTCCCGAGACGGAAGAGAACGTGCTGTGGCAGCTGTTCGGGCCCTTCGGCGCGGTGCAGAGCGTGAAAGTGATCCGCGACCTGCAGACGAACAAGTGCAAGGGATACGGGTTCATCACGATGACCAACTACGACGAGGCTGTGGTCGCCATCCAGTCCCTGAACGGGTACACGCTCGGTAACAGAGTGCTGCAAGTCAGCTTCAAGACCAACAAGATAAAGACGATCTAA
Protein
MSKGDSEQQNGSGEESKTNLIINYLPQSMTQEEIRSLFSSIGEVESCKLIRNKGAAFPDALNHALHGGGQSLGYAFVNYHRPEDAEKAIATLNGLRLQNKTIKVSYARPSSEAIKGANLYVSGLPKTMTQSELERLFSPYGRIITSRILCENSGGRPFTGGEQGLSKGVGFIRFDQRVEAERAIQELNGTIPKGASEPITVKFANNPSNNGKALAPLAAYLPAALRFPAPLGRFSSGKSLLAINKGLQRYSPLAGELLGGVLPGAVGSEWCIFVYNLAPETEENVLWQLFGPFGAVQSVKVIRDLQTNKCKGYGFITMTNYDEAVVAIQSLNGYTLGNRVLQVSFKTNKIKTI
Summary
Uniprot
H9JWA9
A0A0N1PHT7
A0A194QEQ0
A0A2A4JZ37
A0A2H1VP98
A0A0L7LH58
+ More
A0A212F1R7 A0A087ZTS9 A0A0L7RB98 A0A195D982 A0A023F5T6 A0A026WCC9 E2C5I2 A0A1B6LEB4 A0A1B6L707 A0A1B6MC15 A0A1B6EUI0 A0A1B6GJH1 A0A1B6IXA5 A0A1B6CCD8 A0A1B6FTM9 A0A1B6JXI7 A0A1B6K286 A0A1B6DPH5 A0A2H8TD63 A0A0A9XNL4 A0A151X4H5 A0A0A9WQK4 A0A195BR83 W8B765 V9I6M9 A0A0L0C3B0 A0A154PLR5 A0A067R4A9 V9I8A0 V9I6N2 A0A1I8Q1J1 A0A0Q9W7Y3 A0A1B0CNX6 V9I7T6 A0A0Q9XPV3 S6BW20 A0A1B0A194 A0A1I8Q1L8 A0A2A3EDH0 A0A0Q9XF76 A0A1A9YQG7 A0A0K8TL76 A0A034V4J7 K7IPW8 W8AKC4 A0A2R7WD77 A0A087SUZ8 A0A0A1WSN6 A0A0A1XJ57 A0A3B0KLA2 B5DNV4 B3MS25 A0A1S4FEF6 A0A061QLL4 A0A1I8MKP7 A0A1I8Q1M4 B4L604 A0A1W4VJY2 B4M748 B4Q1Z0 A0A0M4EU27 B4NEI3 B4JM01 B3NX84 E1NZB4 Q175F4 A0A1I8MKP3 Q16YJ6 A0A1I8Q1K5 A0A1Q3FNT3 A0A1Q3FNK7 A0A182H8F0 A0A023ERC3 A0A2M4AVH8 A0A182T4S0 A0A182RRC9 A0A3B0KRV0 A0A0R3P1Z1 A0A182KZ02 A0A182HRD2 A0A0N8NZT3 A0A2L2Y0N9 W5J3C3 A0A2L2Y3M6 A0A2M4BS40 A0A0Q9WVT0 A0A1W4VKQ0 A0A0Q5TKR8 Q9VYI0 A0A0R1EAH8 A0A1E1X282 A0A0Q9XEN8 A0A182IP13 B4IGK8
A0A212F1R7 A0A087ZTS9 A0A0L7RB98 A0A195D982 A0A023F5T6 A0A026WCC9 E2C5I2 A0A1B6LEB4 A0A1B6L707 A0A1B6MC15 A0A1B6EUI0 A0A1B6GJH1 A0A1B6IXA5 A0A1B6CCD8 A0A1B6FTM9 A0A1B6JXI7 A0A1B6K286 A0A1B6DPH5 A0A2H8TD63 A0A0A9XNL4 A0A151X4H5 A0A0A9WQK4 A0A195BR83 W8B765 V9I6M9 A0A0L0C3B0 A0A154PLR5 A0A067R4A9 V9I8A0 V9I6N2 A0A1I8Q1J1 A0A0Q9W7Y3 A0A1B0CNX6 V9I7T6 A0A0Q9XPV3 S6BW20 A0A1B0A194 A0A1I8Q1L8 A0A2A3EDH0 A0A0Q9XF76 A0A1A9YQG7 A0A0K8TL76 A0A034V4J7 K7IPW8 W8AKC4 A0A2R7WD77 A0A087SUZ8 A0A0A1WSN6 A0A0A1XJ57 A0A3B0KLA2 B5DNV4 B3MS25 A0A1S4FEF6 A0A061QLL4 A0A1I8MKP7 A0A1I8Q1M4 B4L604 A0A1W4VJY2 B4M748 B4Q1Z0 A0A0M4EU27 B4NEI3 B4JM01 B3NX84 E1NZB4 Q175F4 A0A1I8MKP3 Q16YJ6 A0A1I8Q1K5 A0A1Q3FNT3 A0A1Q3FNK7 A0A182H8F0 A0A023ERC3 A0A2M4AVH8 A0A182T4S0 A0A182RRC9 A0A3B0KRV0 A0A0R3P1Z1 A0A182KZ02 A0A182HRD2 A0A0N8NZT3 A0A2L2Y0N9 W5J3C3 A0A2L2Y3M6 A0A2M4BS40 A0A0Q9WVT0 A0A1W4VKQ0 A0A0Q5TKR8 Q9VYI0 A0A0R1EAH8 A0A1E1X282 A0A0Q9XEN8 A0A182IP13 B4IGK8
Pubmed
19121390
26354079
26227816
22118469
25474469
24508170
+ More
20798317 25401762 24495485 26108605 24845553 17994087 24382152 26369729 25348373 20075255 25830018 15632085 17510324 25315136 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 24945155 20966253 26561354 20920257 23761445 18057021 12324951 12591606 28503490
20798317 25401762 24495485 26108605 24845553 17994087 24382152 26369729 25348373 20075255 25830018 15632085 17510324 25315136 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 24945155 20966253 26561354 20920257 23761445 18057021 12324951 12591606 28503490
EMBL
BABH01029363
KQ459670
KPJ20486.1
KQ459054
KPJ03899.1
NWSH01000388
+ More
PCG76763.1 ODYU01003631 SOQ42628.1 JTDY01001148 KOB74740.1 AGBW02010820 OWR47685.1 KQ414617 KOC68197.1 KQ981153 KYN08964.1 GBBI01002170 JAC16542.1 KK107274 EZA53725.1 GL452770 EFN76795.1 GEBQ01018133 JAT21844.1 GEBQ01020683 JAT19294.1 GEBQ01006501 JAT33476.1 GECZ01028396 JAS41373.1 GECZ01007219 JAS62550.1 GECU01016121 JAS91585.1 GEDC01026181 GEDC01019872 JAS11117.1 JAS17426.1 GECZ01016221 JAS53548.1 GECU01003814 JAT03893.1 GECU01002152 JAT05555.1 GEDC01009775 JAS27523.1 GFXV01000238 MBW12043.1 GBHO01021277 JAG22327.1 KQ982548 KYQ55271.1 GBHO01033903 JAG09701.1 KQ976424 KYM88469.1 GAMC01017484 JAB89071.1 JR035790 JR035791 JR035792 JR035793 AEY56682.1 AEY56683.1 AEY56684.1 AEY56685.1 JRES01000955 KNC26820.1 KQ434977 KZC12821.1 KK852708 KDR18000.1 JR035794 JR035797 JR035798 JR035799 AEY56686.1 AEY56689.1 AEY56690.1 AEY56691.1 JR035795 JR035796 JR035800 JR035801 AEY56687.1 AEY56688.1 AEY56692.1 AEY56693.1 CH940653 KRF80738.1 AJWK01021069 AJWK01021070 JR035802 JR035803 JR035804 JR035805 AEY56694.1 AEY56695.1 AEY56696.1 AEY56697.1 CH933812 KRG07038.1 AB829504 AB829505 BAN59994.1 BAN59995.1 KZ288291 PBC29216.1 KRG07037.1 GDAI01002687 JAI14916.1 GAKP01021503 GAKP01021499 JAC37449.1 GAMC01017485 GAMC01017483 JAB89070.1 KK854488 PTY16275.1 KK112080 KFM56687.1 GBXI01012767 JAD01525.1 GBXI01003305 JAD10987.1 OUUW01000021 SPP89390.1 CH379065 EDY73206.1 CH902622 EDV34580.1 GBFC01000013 JAC59325.1 EDW05800.1 EDW62615.1 CM000162 EDX01511.1 CP012528 ALC48510.1 CH964239 EDW82152.1 CH916371 EDV91762.1 CH954180 EDV47256.1 BT125663 AE014298 ADN43900.1 AFH07361.1 AFH07363.2 AHN59643.1 CH477401 EAT41715.1 CH477515 EAT39711.1 GFDL01005902 JAV29143.1 GFDL01005898 JAV29147.1 JXUM01029111 KQ560843 KXJ80650.1 GAPW01001885 JAC11713.1 GGFK01011478 MBW44799.1 SPP89389.1 KRT07226.1 APCN01001689 KPU75204.1 IAAA01006536 LAA01731.1 ADMH02002138 ETN58341.1 IAAA01006537 LAA01735.1 GGFJ01006756 MBW55897.1 KRF99576.1 KQS30422.1 KQS30423.1 AF239667 BT120384 AAF43091.1 AAF48215.3 ABW09403.1 ADD16464.1 AFH07360.1 AFH07362.1 AGB95339.1 AGB95340.1 KRK06190.1 KRK06191.1 GFAC01005846 JAT93342.1 KRG07039.1 CH480836 EDW48958.1
PCG76763.1 ODYU01003631 SOQ42628.1 JTDY01001148 KOB74740.1 AGBW02010820 OWR47685.1 KQ414617 KOC68197.1 KQ981153 KYN08964.1 GBBI01002170 JAC16542.1 KK107274 EZA53725.1 GL452770 EFN76795.1 GEBQ01018133 JAT21844.1 GEBQ01020683 JAT19294.1 GEBQ01006501 JAT33476.1 GECZ01028396 JAS41373.1 GECZ01007219 JAS62550.1 GECU01016121 JAS91585.1 GEDC01026181 GEDC01019872 JAS11117.1 JAS17426.1 GECZ01016221 JAS53548.1 GECU01003814 JAT03893.1 GECU01002152 JAT05555.1 GEDC01009775 JAS27523.1 GFXV01000238 MBW12043.1 GBHO01021277 JAG22327.1 KQ982548 KYQ55271.1 GBHO01033903 JAG09701.1 KQ976424 KYM88469.1 GAMC01017484 JAB89071.1 JR035790 JR035791 JR035792 JR035793 AEY56682.1 AEY56683.1 AEY56684.1 AEY56685.1 JRES01000955 KNC26820.1 KQ434977 KZC12821.1 KK852708 KDR18000.1 JR035794 JR035797 JR035798 JR035799 AEY56686.1 AEY56689.1 AEY56690.1 AEY56691.1 JR035795 JR035796 JR035800 JR035801 AEY56687.1 AEY56688.1 AEY56692.1 AEY56693.1 CH940653 KRF80738.1 AJWK01021069 AJWK01021070 JR035802 JR035803 JR035804 JR035805 AEY56694.1 AEY56695.1 AEY56696.1 AEY56697.1 CH933812 KRG07038.1 AB829504 AB829505 BAN59994.1 BAN59995.1 KZ288291 PBC29216.1 KRG07037.1 GDAI01002687 JAI14916.1 GAKP01021503 GAKP01021499 JAC37449.1 GAMC01017485 GAMC01017483 JAB89070.1 KK854488 PTY16275.1 KK112080 KFM56687.1 GBXI01012767 JAD01525.1 GBXI01003305 JAD10987.1 OUUW01000021 SPP89390.1 CH379065 EDY73206.1 CH902622 EDV34580.1 GBFC01000013 JAC59325.1 EDW05800.1 EDW62615.1 CM000162 EDX01511.1 CP012528 ALC48510.1 CH964239 EDW82152.1 CH916371 EDV91762.1 CH954180 EDV47256.1 BT125663 AE014298 ADN43900.1 AFH07361.1 AFH07363.2 AHN59643.1 CH477401 EAT41715.1 CH477515 EAT39711.1 GFDL01005902 JAV29143.1 GFDL01005898 JAV29147.1 JXUM01029111 KQ560843 KXJ80650.1 GAPW01001885 JAC11713.1 GGFK01011478 MBW44799.1 SPP89389.1 KRT07226.1 APCN01001689 KPU75204.1 IAAA01006536 LAA01731.1 ADMH02002138 ETN58341.1 IAAA01006537 LAA01735.1 GGFJ01006756 MBW55897.1 KRF99576.1 KQS30422.1 KQS30423.1 AF239667 BT120384 AAF43091.1 AAF48215.3 ABW09403.1 ADD16464.1 AFH07360.1 AFH07362.1 AGB95339.1 AGB95340.1 KRK06190.1 KRK06191.1 GFAC01005846 JAT93342.1 KRG07039.1 CH480836 EDW48958.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000037510
UP000007151
+ More
UP000005203 UP000053825 UP000078492 UP000053097 UP000008237 UP000075809 UP000078540 UP000037069 UP000076502 UP000027135 UP000095300 UP000008792 UP000092461 UP000009192 UP000092445 UP000242457 UP000092443 UP000002358 UP000054359 UP000268350 UP000001819 UP000007801 UP000095301 UP000192221 UP000002282 UP000092553 UP000007798 UP000001070 UP000008711 UP000000803 UP000008820 UP000069940 UP000249989 UP000075901 UP000075900 UP000075882 UP000075840 UP000000673 UP000075880 UP000001292
UP000005203 UP000053825 UP000078492 UP000053097 UP000008237 UP000075809 UP000078540 UP000037069 UP000076502 UP000027135 UP000095300 UP000008792 UP000092461 UP000009192 UP000092445 UP000242457 UP000092443 UP000002358 UP000054359 UP000268350 UP000001819 UP000007801 UP000095301 UP000192221 UP000002282 UP000092553 UP000007798 UP000001070 UP000008711 UP000000803 UP000008820 UP000069940 UP000249989 UP000075901 UP000075900 UP000075882 UP000075840 UP000000673 UP000075880 UP000001292
Pfam
PF00076 RRM_1
Interpro
SUPFAM
SSF54928
SSF54928
Gene 3D
CDD
ProteinModelPortal
H9JWA9
A0A0N1PHT7
A0A194QEQ0
A0A2A4JZ37
A0A2H1VP98
A0A0L7LH58
+ More
A0A212F1R7 A0A087ZTS9 A0A0L7RB98 A0A195D982 A0A023F5T6 A0A026WCC9 E2C5I2 A0A1B6LEB4 A0A1B6L707 A0A1B6MC15 A0A1B6EUI0 A0A1B6GJH1 A0A1B6IXA5 A0A1B6CCD8 A0A1B6FTM9 A0A1B6JXI7 A0A1B6K286 A0A1B6DPH5 A0A2H8TD63 A0A0A9XNL4 A0A151X4H5 A0A0A9WQK4 A0A195BR83 W8B765 V9I6M9 A0A0L0C3B0 A0A154PLR5 A0A067R4A9 V9I8A0 V9I6N2 A0A1I8Q1J1 A0A0Q9W7Y3 A0A1B0CNX6 V9I7T6 A0A0Q9XPV3 S6BW20 A0A1B0A194 A0A1I8Q1L8 A0A2A3EDH0 A0A0Q9XF76 A0A1A9YQG7 A0A0K8TL76 A0A034V4J7 K7IPW8 W8AKC4 A0A2R7WD77 A0A087SUZ8 A0A0A1WSN6 A0A0A1XJ57 A0A3B0KLA2 B5DNV4 B3MS25 A0A1S4FEF6 A0A061QLL4 A0A1I8MKP7 A0A1I8Q1M4 B4L604 A0A1W4VJY2 B4M748 B4Q1Z0 A0A0M4EU27 B4NEI3 B4JM01 B3NX84 E1NZB4 Q175F4 A0A1I8MKP3 Q16YJ6 A0A1I8Q1K5 A0A1Q3FNT3 A0A1Q3FNK7 A0A182H8F0 A0A023ERC3 A0A2M4AVH8 A0A182T4S0 A0A182RRC9 A0A3B0KRV0 A0A0R3P1Z1 A0A182KZ02 A0A182HRD2 A0A0N8NZT3 A0A2L2Y0N9 W5J3C3 A0A2L2Y3M6 A0A2M4BS40 A0A0Q9WVT0 A0A1W4VKQ0 A0A0Q5TKR8 Q9VYI0 A0A0R1EAH8 A0A1E1X282 A0A0Q9XEN8 A0A182IP13 B4IGK8
A0A212F1R7 A0A087ZTS9 A0A0L7RB98 A0A195D982 A0A023F5T6 A0A026WCC9 E2C5I2 A0A1B6LEB4 A0A1B6L707 A0A1B6MC15 A0A1B6EUI0 A0A1B6GJH1 A0A1B6IXA5 A0A1B6CCD8 A0A1B6FTM9 A0A1B6JXI7 A0A1B6K286 A0A1B6DPH5 A0A2H8TD63 A0A0A9XNL4 A0A151X4H5 A0A0A9WQK4 A0A195BR83 W8B765 V9I6M9 A0A0L0C3B0 A0A154PLR5 A0A067R4A9 V9I8A0 V9I6N2 A0A1I8Q1J1 A0A0Q9W7Y3 A0A1B0CNX6 V9I7T6 A0A0Q9XPV3 S6BW20 A0A1B0A194 A0A1I8Q1L8 A0A2A3EDH0 A0A0Q9XF76 A0A1A9YQG7 A0A0K8TL76 A0A034V4J7 K7IPW8 W8AKC4 A0A2R7WD77 A0A087SUZ8 A0A0A1WSN6 A0A0A1XJ57 A0A3B0KLA2 B5DNV4 B3MS25 A0A1S4FEF6 A0A061QLL4 A0A1I8MKP7 A0A1I8Q1M4 B4L604 A0A1W4VJY2 B4M748 B4Q1Z0 A0A0M4EU27 B4NEI3 B4JM01 B3NX84 E1NZB4 Q175F4 A0A1I8MKP3 Q16YJ6 A0A1I8Q1K5 A0A1Q3FNT3 A0A1Q3FNK7 A0A182H8F0 A0A023ERC3 A0A2M4AVH8 A0A182T4S0 A0A182RRC9 A0A3B0KRV0 A0A0R3P1Z1 A0A182KZ02 A0A182HRD2 A0A0N8NZT3 A0A2L2Y0N9 W5J3C3 A0A2L2Y3M6 A0A2M4BS40 A0A0Q9WVT0 A0A1W4VKQ0 A0A0Q5TKR8 Q9VYI0 A0A0R1EAH8 A0A1E1X282 A0A0Q9XEN8 A0A182IP13 B4IGK8
PDB
1FNX
E-value=4.69136e-63,
Score=611
Ontologies
GO
Topology
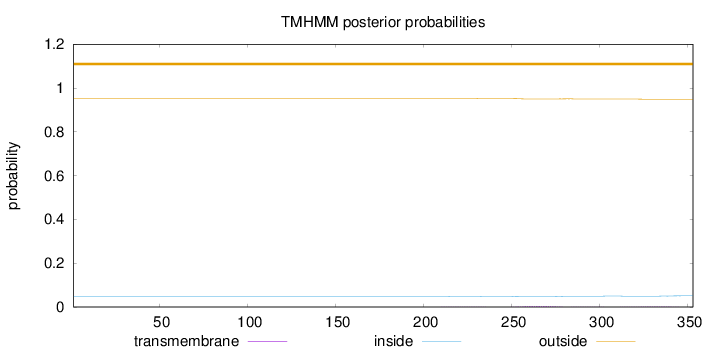
Length:
353
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15051
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.04825
outside
1 - 353
Population Genetic Test Statistics
Pi
258.7311
Theta
208.725356
Tajima's D
0.774788
CLR
0
CSRT
0.598670066496675
Interpretation
Uncertain
