Gene
KWMTBOMO16432
Pre Gene Modal
BGIBMGA013870
Annotation
PREDICTED:_checkpoint_protein_HUS1_[Amyelois_transitella]
Full name
Checkpoint protein
Location in the cell
Nuclear Reliability : 3.127
Sequence
CDS
ATGAAATTCCGTGCTGTTATGAATAACTCTTCTCCAATACGCGAATTCTCAACTATTGTTAGTACAATATCAAAACTGTCTAAGGAGTGTGTCCTAAGGATAGCAGACGATATGTTGTATTTCATTGTCAGCGATGAAAACACTGGACCAGCCCCTCCGGTACTATGGTGTGAAATACCACAAAGTGTATTTTTCTCTGAATATCAATTGGTTGGAATGGATGAAACTCACCAAGACATTTATTTAGGATTGAATTCGGGTAATCTCGCAAGATCTTTTGTCTCCTTGAAAAATGTAAAATCATTAAAAATGAAGTTAACAAAGAAACAGTGTCCGTGCCTAACATTGGAAATTGAAGTGCCGTCTACAACCTCTCATCAGACGAGGCAGATTGTGCATGACATTCCAGTAGCAGTTGTGCCATGGAAAAACTGGCCAGACTTTTATGAACCCAGAGTACCAGATCCTGATATATCAATAGAATTACCAAGTTTGAAACAGCTACGTACAACAATAGATCGTATGAGAACGATGGCATCTGAGGTGATGGTGCGAGCTTCTGCTGGTGGTCAATTAATCTTACAGATCAGAGCAGACATGGCTAAAGTGTCTACAAGGTTTAAAGATTTGAATGTTGCTCATTTTGATGGCCCAGTAGACAATTCGGACCCTGAAACGGACAGCCAAGGAAATGAAGATAGCTCAGAAATAGTACAGTGTCGTGTTGACGCAAAGAAACTAGCACTTTTCTTGAACGTTGATCACATATCAACGAATACAACATTATGTAGTATTGTGCACAAGAGACTTGTGACATTGTGCTTACAGACAGATGATAATGTTAAATTGCAATGTTTCATAAGTGGAATTGTGTATTGA
Protein
MKFRAVMNNSSPIREFSTIVSTISKLSKECVLRIADDMLYFIVSDENTGPAPPVLWCEIPQSVFFSEYQLVGMDETHQDIYLGLNSGNLARSFVSLKNVKSLKMKLTKKQCPCLTLEIEVPSTTSHQTRQIVHDIPVAVVPWKNWPDFYEPRVPDPDISIELPSLKQLRTTIDRMRTMASEVMVRASAGGQLILQIRADMAKVSTRFKDLNVAHFDGPVDNSDPETDSQGNEDSSEIVQCRVDAKKLALFLNVDHISTNTTLCSIVHKRLVTLCLQTDDNVKLQCFISGIVY
Summary
Similarity
Belongs to the HUS1 family.
Uniprot
A0A2H1W8Y3
A0A1E1WPV9
A0A2A4K3P6
A0A3S2TGQ6
A0A2W1BLW8
A0A194Q4R4
+ More
A0A2A4K2N8 S4PT77 A0A139WEK8 A0A1Y1LJB5 A0A0T6BEM5 F7AYQ2 A0A2B4SVH5 A0A1W4Y5B7 A0A098LZL1 A0A3B3R9R7 U3KFE5 W5L6K6 A0A151PCD2 A0A0Q3MEQ3 A0A3S1BK90 A0A3M6UFR5 A0A1A8LQJ8 A0A060XAV8 W5U8V2 A0A2Z5TRN6 A0A3B4CDW3 A0A099Z7V2 A0A1S3QAE5 A0A3P8YK30 A0A3Q1DG78 A0A3P8RV68 C1BF59 A0A3Q3N2E2 G3W0W1 A0A0K8S0G0 A0A1V4JV92 A0A1A8EHR6 A0A3L8SLR7 A0A091K5E9 A0A067RHI8 A0A1A8V9Z7 H2LEI5 A0A3B4UXJ8 A0A3Q3N5W5 A0A1U8CWX9 A0A3P9HEW8 A0A091JVR0 A0A1A8F283 A0A3B4XGF5 A0A3Q4BUI4 A0A3B4YXS1 A0A3Q1IT57 A0A093PPC0 A0A3P9KM55 A0A2I0M4W8 G1KHF0 A0A218UHG1 A0A0B6Z858 A0A1A8PBQ3 A0A1A8IZC5 A0A3Q2XZD9 G1MR80 A0A093I4W5 A3KP28 A0A3B3RAJ3 A0A3B3B5Y9 H0Z9Q2 A0A3S2M314 A0A1D5PTJ5 A0A3P8WSG9 A0A1S3I6X9 A0A091QI02 A0A2I4ALZ8 A0A0L8HS17 A0A0F8CFC5 A0A1A7X915 I3K6T5 A0A1S3H1U6 A0A3Q3N2B7 A0A3Q3AQ45 A0A091FTS6 Q7ZTA4 A0A2J7QDP9 A0A3Q2WLW7 A0A3P8QCN7 A0A3P9CXW6 A0A091NJS8 A7RGK3 A0A026WDN0 A0A091L9V1 U3IFN8 N6TU68 I3MYM9 R0LKE3 A0A091VCS7
A0A2A4K2N8 S4PT77 A0A139WEK8 A0A1Y1LJB5 A0A0T6BEM5 F7AYQ2 A0A2B4SVH5 A0A1W4Y5B7 A0A098LZL1 A0A3B3R9R7 U3KFE5 W5L6K6 A0A151PCD2 A0A0Q3MEQ3 A0A3S1BK90 A0A3M6UFR5 A0A1A8LQJ8 A0A060XAV8 W5U8V2 A0A2Z5TRN6 A0A3B4CDW3 A0A099Z7V2 A0A1S3QAE5 A0A3P8YK30 A0A3Q1DG78 A0A3P8RV68 C1BF59 A0A3Q3N2E2 G3W0W1 A0A0K8S0G0 A0A1V4JV92 A0A1A8EHR6 A0A3L8SLR7 A0A091K5E9 A0A067RHI8 A0A1A8V9Z7 H2LEI5 A0A3B4UXJ8 A0A3Q3N5W5 A0A1U8CWX9 A0A3P9HEW8 A0A091JVR0 A0A1A8F283 A0A3B4XGF5 A0A3Q4BUI4 A0A3B4YXS1 A0A3Q1IT57 A0A093PPC0 A0A3P9KM55 A0A2I0M4W8 G1KHF0 A0A218UHG1 A0A0B6Z858 A0A1A8PBQ3 A0A1A8IZC5 A0A3Q2XZD9 G1MR80 A0A093I4W5 A3KP28 A0A3B3RAJ3 A0A3B3B5Y9 H0Z9Q2 A0A3S2M314 A0A1D5PTJ5 A0A3P8WSG9 A0A1S3I6X9 A0A091QI02 A0A2I4ALZ8 A0A0L8HS17 A0A0F8CFC5 A0A1A7X915 I3K6T5 A0A1S3H1U6 A0A3Q3N2B7 A0A3Q3AQ45 A0A091FTS6 Q7ZTA4 A0A2J7QDP9 A0A3Q2WLW7 A0A3P8QCN7 A0A3P9CXW6 A0A091NJS8 A7RGK3 A0A026WDN0 A0A091L9V1 U3IFN8 N6TU68 I3MYM9 R0LKE3 A0A091VCS7
Pubmed
28756777
26354079
23622113
18362917
19820115
28004739
+ More
17495919 29240929 25329095 22293439 30382153 24755649 23127152 26760975 25069045 21709235 25727380 30282656 24845553 17554307 23371554 20838655 29451363 20360741 15592404 24487278 25835551 25186727 12620222 17615350 24508170 23749191 23537049
17495919 29240929 25329095 22293439 30382153 24755649 23127152 26760975 25069045 21709235 25727380 30282656 24845553 17554307 23371554 20838655 29451363 20360741 15592404 24487278 25835551 25186727 12620222 17615350 24508170 23749191 23537049
EMBL
ODYU01007096
SOQ49559.1
GDQN01002183
JAT88871.1
NWSH01000196
PCG78514.1
+ More
RSAL01000157 RVE45561.1 KZ150002 PZC75281.1 KQ459463 KPJ00528.1 PCG78515.1 GAIX01012518 JAA80042.1 KQ971354 KYB26297.1 GEZM01057094 JAV72450.1 LJIG01001165 KRT85775.1 LSMT01000008 PFX33881.1 GBSI01001121 JAC95375.1 AGTO01011182 AKHW03000499 KYO46584.1 LMAW01002505 KQK80840.1 RQTK01000161 RUS85704.1 RCHS01001620 RMX52527.1 HAEF01008429 HAEG01004377 SBR46586.1 FR905132 CDQ76362.1 JT408086 JT413130 AHH38230.1 FX985750 BBA93637.1 KL889742 KGL77138.1 BT073238 ACO07662.1 AEFK01086447 AEFK01086448 GBKD01001009 JAG46609.1 LSYS01006073 OPJ76108.1 HADZ01000576 HAEA01017222 SBQ45703.1 QUSF01000013 RLW04360.1 KK543368 KFP31541.1 KK852680 KDR18613.1 HADY01004146 HAEJ01016728 SBS57185.1 KK501502 KFP15656.1 HAEB01006918 HAEC01016261 SBQ53445.1 KL670090 KFW78291.1 AKCR02000040 PKK24720.1 MUZQ01000301 OWK53193.1 HACG01017848 CEK64713.1 HAEH01006245 SBR78706.1 HAED01016290 SBR02735.1 KL206835 KFV86252.1 BC134133 AAI34134.1 ABQF01023845 ABQF01023846 CM012456 RVE59094.1 AC145931 KK662511 KFQ09104.1 KQ417429 KOF92006.1 KQ041022 KKF31417.1 HADW01013197 SBP14597.1 AERX01028249 KK719661 KFO64590.1 AY169966 AAO13094.1 NEVH01015817 PNF26711.1 KL385255 KFP89696.1 DS469509 EDO49542.1 KK107260 EZA54157.1 KL311152 KFP53144.1 ADON01064265 APGK01052269 KB741211 ENN72820.1 AGTP01012998 KB742991 EOB02150.1 KL410864 KFR00218.1
RSAL01000157 RVE45561.1 KZ150002 PZC75281.1 KQ459463 KPJ00528.1 PCG78515.1 GAIX01012518 JAA80042.1 KQ971354 KYB26297.1 GEZM01057094 JAV72450.1 LJIG01001165 KRT85775.1 LSMT01000008 PFX33881.1 GBSI01001121 JAC95375.1 AGTO01011182 AKHW03000499 KYO46584.1 LMAW01002505 KQK80840.1 RQTK01000161 RUS85704.1 RCHS01001620 RMX52527.1 HAEF01008429 HAEG01004377 SBR46586.1 FR905132 CDQ76362.1 JT408086 JT413130 AHH38230.1 FX985750 BBA93637.1 KL889742 KGL77138.1 BT073238 ACO07662.1 AEFK01086447 AEFK01086448 GBKD01001009 JAG46609.1 LSYS01006073 OPJ76108.1 HADZ01000576 HAEA01017222 SBQ45703.1 QUSF01000013 RLW04360.1 KK543368 KFP31541.1 KK852680 KDR18613.1 HADY01004146 HAEJ01016728 SBS57185.1 KK501502 KFP15656.1 HAEB01006918 HAEC01016261 SBQ53445.1 KL670090 KFW78291.1 AKCR02000040 PKK24720.1 MUZQ01000301 OWK53193.1 HACG01017848 CEK64713.1 HAEH01006245 SBR78706.1 HAED01016290 SBR02735.1 KL206835 KFV86252.1 BC134133 AAI34134.1 ABQF01023845 ABQF01023846 CM012456 RVE59094.1 AC145931 KK662511 KFQ09104.1 KQ417429 KOF92006.1 KQ041022 KKF31417.1 HADW01013197 SBP14597.1 AERX01028249 KK719661 KFO64590.1 AY169966 AAO13094.1 NEVH01015817 PNF26711.1 KL385255 KFP89696.1 DS469509 EDO49542.1 KK107260 EZA54157.1 KL311152 KFP53144.1 ADON01064265 APGK01052269 KB741211 ENN72820.1 AGTP01012998 KB742991 EOB02150.1 KL410864 KFR00218.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000007266
UP000002280
UP000225706
+ More
UP000192224 UP000261540 UP000016665 UP000018467 UP000050525 UP000051836 UP000271974 UP000275408 UP000193380 UP000221080 UP000261440 UP000053641 UP000087266 UP000265140 UP000257160 UP000265080 UP000261660 UP000007648 UP000190648 UP000276834 UP000027135 UP000001038 UP000261420 UP000261640 UP000189705 UP000265200 UP000053119 UP000261360 UP000261620 UP000261400 UP000265040 UP000053258 UP000265180 UP000053872 UP000001646 UP000197619 UP000264820 UP000001645 UP000053584 UP000261560 UP000007754 UP000000539 UP000265120 UP000085678 UP000192220 UP000053454 UP000005207 UP000264800 UP000052976 UP000235965 UP000264840 UP000265100 UP000265160 UP000001593 UP000053097 UP000016666 UP000019118 UP000005215 UP000053283
UP000192224 UP000261540 UP000016665 UP000018467 UP000050525 UP000051836 UP000271974 UP000275408 UP000193380 UP000221080 UP000261440 UP000053641 UP000087266 UP000265140 UP000257160 UP000265080 UP000261660 UP000007648 UP000190648 UP000276834 UP000027135 UP000001038 UP000261420 UP000261640 UP000189705 UP000265200 UP000053119 UP000261360 UP000261620 UP000261400 UP000265040 UP000053258 UP000265180 UP000053872 UP000001646 UP000197619 UP000264820 UP000001645 UP000053584 UP000261560 UP000007754 UP000000539 UP000265120 UP000085678 UP000192220 UP000053454 UP000005207 UP000264800 UP000052976 UP000235965 UP000264840 UP000265100 UP000265160 UP000001593 UP000053097 UP000016666 UP000019118 UP000005215 UP000053283
Pfam
PF04005 Hus1
ProteinModelPortal
A0A2H1W8Y3
A0A1E1WPV9
A0A2A4K3P6
A0A3S2TGQ6
A0A2W1BLW8
A0A194Q4R4
+ More
A0A2A4K2N8 S4PT77 A0A139WEK8 A0A1Y1LJB5 A0A0T6BEM5 F7AYQ2 A0A2B4SVH5 A0A1W4Y5B7 A0A098LZL1 A0A3B3R9R7 U3KFE5 W5L6K6 A0A151PCD2 A0A0Q3MEQ3 A0A3S1BK90 A0A3M6UFR5 A0A1A8LQJ8 A0A060XAV8 W5U8V2 A0A2Z5TRN6 A0A3B4CDW3 A0A099Z7V2 A0A1S3QAE5 A0A3P8YK30 A0A3Q1DG78 A0A3P8RV68 C1BF59 A0A3Q3N2E2 G3W0W1 A0A0K8S0G0 A0A1V4JV92 A0A1A8EHR6 A0A3L8SLR7 A0A091K5E9 A0A067RHI8 A0A1A8V9Z7 H2LEI5 A0A3B4UXJ8 A0A3Q3N5W5 A0A1U8CWX9 A0A3P9HEW8 A0A091JVR0 A0A1A8F283 A0A3B4XGF5 A0A3Q4BUI4 A0A3B4YXS1 A0A3Q1IT57 A0A093PPC0 A0A3P9KM55 A0A2I0M4W8 G1KHF0 A0A218UHG1 A0A0B6Z858 A0A1A8PBQ3 A0A1A8IZC5 A0A3Q2XZD9 G1MR80 A0A093I4W5 A3KP28 A0A3B3RAJ3 A0A3B3B5Y9 H0Z9Q2 A0A3S2M314 A0A1D5PTJ5 A0A3P8WSG9 A0A1S3I6X9 A0A091QI02 A0A2I4ALZ8 A0A0L8HS17 A0A0F8CFC5 A0A1A7X915 I3K6T5 A0A1S3H1U6 A0A3Q3N2B7 A0A3Q3AQ45 A0A091FTS6 Q7ZTA4 A0A2J7QDP9 A0A3Q2WLW7 A0A3P8QCN7 A0A3P9CXW6 A0A091NJS8 A7RGK3 A0A026WDN0 A0A091L9V1 U3IFN8 N6TU68 I3MYM9 R0LKE3 A0A091VCS7
A0A2A4K2N8 S4PT77 A0A139WEK8 A0A1Y1LJB5 A0A0T6BEM5 F7AYQ2 A0A2B4SVH5 A0A1W4Y5B7 A0A098LZL1 A0A3B3R9R7 U3KFE5 W5L6K6 A0A151PCD2 A0A0Q3MEQ3 A0A3S1BK90 A0A3M6UFR5 A0A1A8LQJ8 A0A060XAV8 W5U8V2 A0A2Z5TRN6 A0A3B4CDW3 A0A099Z7V2 A0A1S3QAE5 A0A3P8YK30 A0A3Q1DG78 A0A3P8RV68 C1BF59 A0A3Q3N2E2 G3W0W1 A0A0K8S0G0 A0A1V4JV92 A0A1A8EHR6 A0A3L8SLR7 A0A091K5E9 A0A067RHI8 A0A1A8V9Z7 H2LEI5 A0A3B4UXJ8 A0A3Q3N5W5 A0A1U8CWX9 A0A3P9HEW8 A0A091JVR0 A0A1A8F283 A0A3B4XGF5 A0A3Q4BUI4 A0A3B4YXS1 A0A3Q1IT57 A0A093PPC0 A0A3P9KM55 A0A2I0M4W8 G1KHF0 A0A218UHG1 A0A0B6Z858 A0A1A8PBQ3 A0A1A8IZC5 A0A3Q2XZD9 G1MR80 A0A093I4W5 A3KP28 A0A3B3RAJ3 A0A3B3B5Y9 H0Z9Q2 A0A3S2M314 A0A1D5PTJ5 A0A3P8WSG9 A0A1S3I6X9 A0A091QI02 A0A2I4ALZ8 A0A0L8HS17 A0A0F8CFC5 A0A1A7X915 I3K6T5 A0A1S3H1U6 A0A3Q3N2B7 A0A3Q3AQ45 A0A091FTS6 Q7ZTA4 A0A2J7QDP9 A0A3Q2WLW7 A0A3P8QCN7 A0A3P9CXW6 A0A091NJS8 A7RGK3 A0A026WDN0 A0A091L9V1 U3IFN8 N6TU68 I3MYM9 R0LKE3 A0A091VCS7
PDB
3G65
E-value=2.52983e-49,
Score=492
Ontologies
GO
Topology
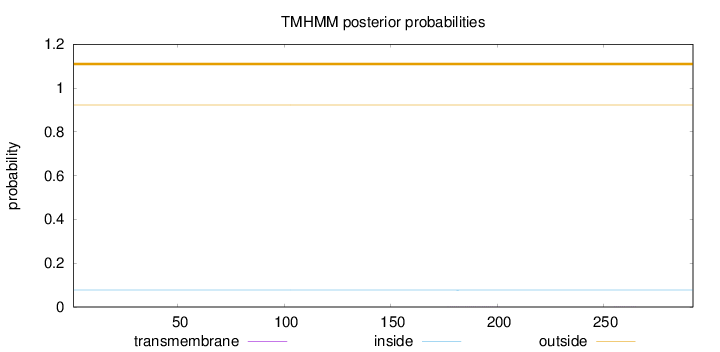
Length:
292
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01505
Exp number, first 60 AAs:
0.000690000000000001
Total prob of N-in:
0.07772
outside
1 - 292
Population Genetic Test Statistics
Pi
312.967613
Theta
182.481871
Tajima's D
2.094245
CLR
0.019648
CSRT
0.895455227238638
Interpretation
Uncertain
