Gene
KWMTBOMO16401
Pre Gene Modal
BGIBMGA013851
Annotation
PREDICTED:_putative_ATP-dependent_RNA_helicase_DHX30_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.181 Mitochondrial Reliability : 1.539
Sequence
CDS
ATGTACATACATTTTATTAACTTTTTTATTTCAGATGATTACTCGGATAATGTTTTCGAAAAGGAAAACAGTACCAACGATGTTTTTGAACAAAAAAACAACGTCCATCCCGTGTACGGAAGGATCATAACGGAGCCGTCGGAAAACACATTAACGAAACGCGAGAGAACTCTGACAAGGAAATTTGAACTTTACGACGCCGAGAAAACTCCTTTGCCCATCGATGGATACGCTAAGCAAATATCTGAGGCGGTCGAAGCCAGCCGGGTGACTGTGGTTGTGGGGGAAGCTGGCTGTGGGAAGTCCACGCGAGTGCCGATCGCGTTGTTGCAAAACTCCTTGTTGTCGACCGCCATTGTGTCGGAGCCGAGACGCGTGGCCGCCATCGGACTGGCGCAGAGAGTCGCCAGCGAACTAGGGGAGGAGGTTGGAGAAACAGTCGGTTATCAAGTGCGCTTGCACTCGAAGCCGCCGCGGCCACCCGCCGGCTCCGTGCTCTACTGCACCAGCGGGGTCCTGCTGAGGAGGCTGCAGGCCAACCCAGGTCTCGTAGGCTGCACTCACGTGCTCATAGATGAGGCTCACGAGAGGGACGTGAACACCGATGTGACCTTATTGCTTTTGAAAAGAGCTTTGAAGATCAATTCGGAGCTTAAAGTCGTTATAATGAGCGCCACGCTTGATGTCGACGTGTTCACTAGATATTTCGGACAATGTCCGGTGATAGCAGTTCCGGGGAGAACTTACCCGGTCGAGGTCACGTATCTCGATGACATACAAAAGAAGTTCAATTTGAATTTGCCCAAAACGATGGAGAGTATCTCGAGGGCGGGCGGTAGACCGACGGTCCACTGCCAAGAGATAGCCGAAGTGATAAAAGCGATCGACAGAACGCAAGAGGAAGGTGCGATTTTAGTGTTCCTCCCGGGCTGGGCCGAGATAAAATATACGATGCAGATATTAGAAGAGCACTATCGAGGCTCGGACTTGCATATGCTCGTGCCTGTTCATTCCAGACTGTCGGCGTTGGAGCAGGCGCGCATATTCTCGGCGGCGGCGGCGGGCGTGCGGCGCGTGCTGCTGGCGACCAACGTGGCGGAGACGTCGCTGACGGTGCCGGACGTCACGCACGTGCTGGACACCGGCGCGCACCGGGAGAACACCACCGACCAGAGCGCGGGTACATTATGTCTGGAGACGGCGTGGGTGTCGCAGGCCGCCGCCAAGCAGAGGTGCGGACGAGCCGGGAGAGTGCGAGCGGGTCACTGCTACAAGCTGTACACGAGGCAGAAGGAGCAGGAGTTCCGGCCGCACTCCACCCCCGAGATACTGAGGGTGCCGCTGGTGCAGACGGTGCTGGACTGCAAGTCGTACGCTCCGGACGACAGGATCGAGGACTTCCTGTCGCAGCTGCCGGAGCCCCCCGGCTCGGACGCTATCGACTTCGCCGTCAACGACCTCGTGGAGATTGGTGCTCTAACTAGCCGGCAACAGCTGACTCGTCTCGGCGGGCTGTTGTGCGGCCTGGCGCTGTCGCCGCGGGTCGCCGTCTGCCTGCTACACTCGGTGGCCATTGGCAATGTCATTGCCGCCGCCACGATCGCCACGCACGTCAGCGACAACCTGCAGCTGTTCCACGACGTTGCTCAACGCAAGGCAGAAATACTCGAGATGAAGCGCAAGTTCAGTCCGACGTGTGACTACAGCGCCCTGTACTGGATCCAGCGCGAGTTCGAACAGAAGCTGTCCGAGCTCGGCGGGGGGGAGGCGGGGCGCTGGTGCACGCAGCACGGACTCAGTAAGGAGAGATTGAAAAACGTTAAAGTGTTTGTTAATAAGCGCGCGGCGTGA
Protein
MYIHFINFFISDDYSDNVFEKENSTNDVFEQKNNVHPVYGRIITEPSENTLTKRERTLTRKFELYDAEKTPLPIDGYAKQISEAVEASRVTVVVGEAGCGKSTRVPIALLQNSLLSTAIVSEPRRVAAIGLAQRVASELGEEVGETVGYQVRLHSKPPRPPAGSVLYCTSGVLLRRLQANPGLVGCTHVLIDEAHERDVNTDVTLLLLKRALKINSELKVVIMSATLDVDVFTRYFGQCPVIAVPGRTYPVEVTYLDDIQKKFNLNLPKTMESISRAGGRPTVHCQEIAEVIKAIDRTQEEGAILVFLPGWAEIKYTMQILEEHYRGSDLHMLVPVHSRLSALEQARIFSAAAAGVRRVLLATNVAETSLTVPDVTHVLDTGAHRENTTDQSAGTLCLETAWVSQAAAKQRCGRAGRVRAGHCYKLYTRQKEQEFRPHSTPEILRVPLVQTVLDCKSYAPDDRIEDFLSQLPEPPGSDAIDFAVNDLVEIGALTSRQQLTRLGGLLCGLALSPRVAVCLLHSVAIGNVIAAATIATHVSDNLQLFHDVAQRKAEILEMKRKFSPTCDYSALYWIQREFEQKLSELGGGEAGRWCTQHGLSKERLKNVKVFVNKRAA
Summary
Uniprot
H9JWD7
A0A2H1W358
A0A2W1BD14
A0A3S2P6Z0
A0A2H1W369
A0A0N1I2R1
+ More
A0A2J7QIM6 A0A2J7QIK8 A0A2J7QIM2 A0A1B6D5F5 A0A1B6E7S0 A0A067R1K9 A0A0J7N443 A0A232F3R9 E2AQI2 K7J1Y4 D6WR01 A0A1B6JUF2 A0A1B6KUX6 A0A0C9RAH5 A0A0P5HD25 A0A0N8CFU7 A0A0P5Q7S4 A0A0P5ZV55 E9FW51 A0A162SS74 A0A0P6HSL9 A0A0P6EBC2 A0A0P5W8I3 A0A3S3PG41 A0A0P5Z7N0 A0A2P8Z378 A0A1W4XGG8 A0A195BRT6 A0A1Y1M4R4 A0A1Y1M7C7 Q4SLJ6 A0A3Q3MLB2 A0A3Q3SC80 A0A1A8MCD6 A0A195EX00 A0A1B6FBX0 A0A1A8LX32 A0A1A8NWU2 A0A1Y1M649 A0A3Q2YBA6 A0A158NNZ5 A0A1A8A2H8 A0A1A8PPH4 A0A1A8I8A4 A0A1A8EQ18 A0A3Q2CIP5 A0A3Q3FMK5 H3CSX9 A0A1A8JQ21 A0A3B4G9U5 A0A0P4YBL8 G3Q113 F4WPH9 A0A1A7ZGX7 A0A0P5Y1Q1 A0A3P9DHI3 A0A0N8DVW8 A0A3B3BFP7 A0A1A8PDE3 A0A3Q3BRL1 A0A3P8S1Q8 A0A3Q1BQF0 A0A3Q1CR53 A0A1A8HIY7 A0A3P8QLQ4 A0A3Q3CKF4 A0A3B4X271 A0A2U9BJ26 A0A3P8VLC7 A0A3Q4MPV5 A0A1A7XCB5 A0A1A7YGG4 A0A1A8J218 A0A1A8UL30 A0A1W0WL65 A0A151WL97 H0YPC0 A0A093FLT7 A0A3P9HNX1 A0A3Q2TSG4 A0A3L8RUM3 I3JTA5 A0A091G9X6 A0A087QJJ0 A0A3Q2TWE1 U3JH43 A0A091FID0 A0A218UGY0 A0A091RQJ4 R0JIJ9 A0A3Q1GXA1 A0A3M0J4J1 H2MS81
A0A2J7QIM6 A0A2J7QIK8 A0A2J7QIM2 A0A1B6D5F5 A0A1B6E7S0 A0A067R1K9 A0A0J7N443 A0A232F3R9 E2AQI2 K7J1Y4 D6WR01 A0A1B6JUF2 A0A1B6KUX6 A0A0C9RAH5 A0A0P5HD25 A0A0N8CFU7 A0A0P5Q7S4 A0A0P5ZV55 E9FW51 A0A162SS74 A0A0P6HSL9 A0A0P6EBC2 A0A0P5W8I3 A0A3S3PG41 A0A0P5Z7N0 A0A2P8Z378 A0A1W4XGG8 A0A195BRT6 A0A1Y1M4R4 A0A1Y1M7C7 Q4SLJ6 A0A3Q3MLB2 A0A3Q3SC80 A0A1A8MCD6 A0A195EX00 A0A1B6FBX0 A0A1A8LX32 A0A1A8NWU2 A0A1Y1M649 A0A3Q2YBA6 A0A158NNZ5 A0A1A8A2H8 A0A1A8PPH4 A0A1A8I8A4 A0A1A8EQ18 A0A3Q2CIP5 A0A3Q3FMK5 H3CSX9 A0A1A8JQ21 A0A3B4G9U5 A0A0P4YBL8 G3Q113 F4WPH9 A0A1A7ZGX7 A0A0P5Y1Q1 A0A3P9DHI3 A0A0N8DVW8 A0A3B3BFP7 A0A1A8PDE3 A0A3Q3BRL1 A0A3P8S1Q8 A0A3Q1BQF0 A0A3Q1CR53 A0A1A8HIY7 A0A3P8QLQ4 A0A3Q3CKF4 A0A3B4X271 A0A2U9BJ26 A0A3P8VLC7 A0A3Q4MPV5 A0A1A7XCB5 A0A1A7YGG4 A0A1A8J218 A0A1A8UL30 A0A1W0WL65 A0A151WL97 H0YPC0 A0A093FLT7 A0A3P9HNX1 A0A3Q2TSG4 A0A3L8RUM3 I3JTA5 A0A091G9X6 A0A087QJJ0 A0A3Q2TWE1 U3JH43 A0A091FID0 A0A218UGY0 A0A091RQJ4 R0JIJ9 A0A3Q1GXA1 A0A3M0J4J1 H2MS81
Pubmed
EMBL
BABH01029202
BABH01029203
BABH01029204
ODYU01006002
SOQ47463.1
KZ150265
+ More
PZC71704.1 RSAL01000008 RVE53921.1 ODYU01006004 SOQ47473.1 KQ459547 KPJ00035.1 NEVH01013589 PNF28423.1 PNF28425.1 PNF28424.1 GEDC01016422 JAS20876.1 GEDC01003306 JAS33992.1 KK852779 KDR16683.1 LBMM01010444 KMQ87455.1 NNAY01001011 OXU25496.1 GL441773 EFN64291.1 KQ971351 EFA06533.2 GECU01030878 GECU01004900 GECU01004084 JAS76828.1 JAT02807.1 JAT03623.1 GEBQ01024923 JAT15054.1 GBYB01005265 JAG75032.1 GDIQ01229480 JAK22245.1 GDIP01135871 JAL67843.1 GDIQ01121021 JAL30705.1 GDIP01250757 GDIP01039046 JAM64669.1 GL732525 EFX88984.1 LRGB01000024 KZS21583.1 GDIQ01022714 JAN72023.1 GDIQ01065994 JAN28743.1 GDIP01089726 JAM13989.1 NCKU01002705 RWS08967.1 GDIP01047730 JAM55985.1 PYGN01000217 PSN50954.1 KQ976417 KYM89742.1 GEZM01041341 JAV80501.1 GEZM01041342 JAV80500.1 CAAE01014556 CAF98486.1 HAEF01013303 SBR54462.1 KQ981953 KYN32409.1 GECZ01022039 JAS47730.1 HAEF01009467 SBR48534.1 HAEG01005185 SBR73498.1 GEZM01041343 JAV80498.1 ADTU01021887 ADTU01021888 HADY01009895 SBP48380.1 HAEI01003035 SBR82902.1 HAED01006842 SBQ92970.1 HAEB01002044 SBQ48571.1 HAEE01002188 SBR22208.1 GDIP01233458 JAI89943.1 GL888245 EGI63892.1 HADY01003597 SBP42082.1 GDIP01065232 JAM38483.1 GDIQ01086724 JAN08013.1 HAEH01006465 SBR79296.1 HAEB01008080 HAEC01015979 SBQ84200.1 CP026249 AWP03993.1 HADW01014064 SBP15464.1 HADX01007415 SBP29647.1 HAED01017209 SBR03654.1 HAEJ01007844 SBS48301.1 MTYJ01000080 OQV15950.1 KQ982971 KYQ48616.1 ABQF01060766 KK636450 KFV58687.1 AADN05000033 QUSF01000203 RLV87417.1 AERX01001030 AERX01001031 KL447876 KFO78146.1 KL225654 KFM01394.1 AGTO01011876 KK719059 KFO61145.1 MUZQ01000313 OWK52916.1 KK929278 KFQ44389.1 KB743777 EOA97100.1 QRBI01000204 RMB93833.1
PZC71704.1 RSAL01000008 RVE53921.1 ODYU01006004 SOQ47473.1 KQ459547 KPJ00035.1 NEVH01013589 PNF28423.1 PNF28425.1 PNF28424.1 GEDC01016422 JAS20876.1 GEDC01003306 JAS33992.1 KK852779 KDR16683.1 LBMM01010444 KMQ87455.1 NNAY01001011 OXU25496.1 GL441773 EFN64291.1 KQ971351 EFA06533.2 GECU01030878 GECU01004900 GECU01004084 JAS76828.1 JAT02807.1 JAT03623.1 GEBQ01024923 JAT15054.1 GBYB01005265 JAG75032.1 GDIQ01229480 JAK22245.1 GDIP01135871 JAL67843.1 GDIQ01121021 JAL30705.1 GDIP01250757 GDIP01039046 JAM64669.1 GL732525 EFX88984.1 LRGB01000024 KZS21583.1 GDIQ01022714 JAN72023.1 GDIQ01065994 JAN28743.1 GDIP01089726 JAM13989.1 NCKU01002705 RWS08967.1 GDIP01047730 JAM55985.1 PYGN01000217 PSN50954.1 KQ976417 KYM89742.1 GEZM01041341 JAV80501.1 GEZM01041342 JAV80500.1 CAAE01014556 CAF98486.1 HAEF01013303 SBR54462.1 KQ981953 KYN32409.1 GECZ01022039 JAS47730.1 HAEF01009467 SBR48534.1 HAEG01005185 SBR73498.1 GEZM01041343 JAV80498.1 ADTU01021887 ADTU01021888 HADY01009895 SBP48380.1 HAEI01003035 SBR82902.1 HAED01006842 SBQ92970.1 HAEB01002044 SBQ48571.1 HAEE01002188 SBR22208.1 GDIP01233458 JAI89943.1 GL888245 EGI63892.1 HADY01003597 SBP42082.1 GDIP01065232 JAM38483.1 GDIQ01086724 JAN08013.1 HAEH01006465 SBR79296.1 HAEB01008080 HAEC01015979 SBQ84200.1 CP026249 AWP03993.1 HADW01014064 SBP15464.1 HADX01007415 SBP29647.1 HAED01017209 SBR03654.1 HAEJ01007844 SBS48301.1 MTYJ01000080 OQV15950.1 KQ982971 KYQ48616.1 ABQF01060766 KK636450 KFV58687.1 AADN05000033 QUSF01000203 RLV87417.1 AERX01001030 AERX01001031 KL447876 KFO78146.1 KL225654 KFM01394.1 AGTO01011876 KK719059 KFO61145.1 MUZQ01000313 OWK52916.1 KK929278 KFQ44389.1 KB743777 EOA97100.1 QRBI01000204 RMB93833.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000235965
UP000027135
UP000036403
+ More
UP000215335 UP000000311 UP000002358 UP000007266 UP000000305 UP000076858 UP000285301 UP000245037 UP000192223 UP000078540 UP000261640 UP000078541 UP000264820 UP000005205 UP000265020 UP000261660 UP000007303 UP000261460 UP000007635 UP000007755 UP000265160 UP000261560 UP000264840 UP000265080 UP000257160 UP000265100 UP000261360 UP000246464 UP000265120 UP000261580 UP000075809 UP000007754 UP000265200 UP000000539 UP000276834 UP000005207 UP000053760 UP000053286 UP000016665 UP000052976 UP000197619 UP000257200 UP000269221 UP000001038
UP000215335 UP000000311 UP000002358 UP000007266 UP000000305 UP000076858 UP000285301 UP000245037 UP000192223 UP000078540 UP000261640 UP000078541 UP000264820 UP000005205 UP000265020 UP000261660 UP000007303 UP000261460 UP000007635 UP000007755 UP000265160 UP000261560 UP000264840 UP000265080 UP000257160 UP000265100 UP000261360 UP000246464 UP000265120 UP000261580 UP000075809 UP000007754 UP000265200 UP000000539 UP000276834 UP000005207 UP000053760 UP000053286 UP000016665 UP000052976 UP000197619 UP000257200 UP000269221 UP000001038
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9JWD7
A0A2H1W358
A0A2W1BD14
A0A3S2P6Z0
A0A2H1W369
A0A0N1I2R1
+ More
A0A2J7QIM6 A0A2J7QIK8 A0A2J7QIM2 A0A1B6D5F5 A0A1B6E7S0 A0A067R1K9 A0A0J7N443 A0A232F3R9 E2AQI2 K7J1Y4 D6WR01 A0A1B6JUF2 A0A1B6KUX6 A0A0C9RAH5 A0A0P5HD25 A0A0N8CFU7 A0A0P5Q7S4 A0A0P5ZV55 E9FW51 A0A162SS74 A0A0P6HSL9 A0A0P6EBC2 A0A0P5W8I3 A0A3S3PG41 A0A0P5Z7N0 A0A2P8Z378 A0A1W4XGG8 A0A195BRT6 A0A1Y1M4R4 A0A1Y1M7C7 Q4SLJ6 A0A3Q3MLB2 A0A3Q3SC80 A0A1A8MCD6 A0A195EX00 A0A1B6FBX0 A0A1A8LX32 A0A1A8NWU2 A0A1Y1M649 A0A3Q2YBA6 A0A158NNZ5 A0A1A8A2H8 A0A1A8PPH4 A0A1A8I8A4 A0A1A8EQ18 A0A3Q2CIP5 A0A3Q3FMK5 H3CSX9 A0A1A8JQ21 A0A3B4G9U5 A0A0P4YBL8 G3Q113 F4WPH9 A0A1A7ZGX7 A0A0P5Y1Q1 A0A3P9DHI3 A0A0N8DVW8 A0A3B3BFP7 A0A1A8PDE3 A0A3Q3BRL1 A0A3P8S1Q8 A0A3Q1BQF0 A0A3Q1CR53 A0A1A8HIY7 A0A3P8QLQ4 A0A3Q3CKF4 A0A3B4X271 A0A2U9BJ26 A0A3P8VLC7 A0A3Q4MPV5 A0A1A7XCB5 A0A1A7YGG4 A0A1A8J218 A0A1A8UL30 A0A1W0WL65 A0A151WL97 H0YPC0 A0A093FLT7 A0A3P9HNX1 A0A3Q2TSG4 A0A3L8RUM3 I3JTA5 A0A091G9X6 A0A087QJJ0 A0A3Q2TWE1 U3JH43 A0A091FID0 A0A218UGY0 A0A091RQJ4 R0JIJ9 A0A3Q1GXA1 A0A3M0J4J1 H2MS81
A0A2J7QIM6 A0A2J7QIK8 A0A2J7QIM2 A0A1B6D5F5 A0A1B6E7S0 A0A067R1K9 A0A0J7N443 A0A232F3R9 E2AQI2 K7J1Y4 D6WR01 A0A1B6JUF2 A0A1B6KUX6 A0A0C9RAH5 A0A0P5HD25 A0A0N8CFU7 A0A0P5Q7S4 A0A0P5ZV55 E9FW51 A0A162SS74 A0A0P6HSL9 A0A0P6EBC2 A0A0P5W8I3 A0A3S3PG41 A0A0P5Z7N0 A0A2P8Z378 A0A1W4XGG8 A0A195BRT6 A0A1Y1M4R4 A0A1Y1M7C7 Q4SLJ6 A0A3Q3MLB2 A0A3Q3SC80 A0A1A8MCD6 A0A195EX00 A0A1B6FBX0 A0A1A8LX32 A0A1A8NWU2 A0A1Y1M649 A0A3Q2YBA6 A0A158NNZ5 A0A1A8A2H8 A0A1A8PPH4 A0A1A8I8A4 A0A1A8EQ18 A0A3Q2CIP5 A0A3Q3FMK5 H3CSX9 A0A1A8JQ21 A0A3B4G9U5 A0A0P4YBL8 G3Q113 F4WPH9 A0A1A7ZGX7 A0A0P5Y1Q1 A0A3P9DHI3 A0A0N8DVW8 A0A3B3BFP7 A0A1A8PDE3 A0A3Q3BRL1 A0A3P8S1Q8 A0A3Q1BQF0 A0A3Q1CR53 A0A1A8HIY7 A0A3P8QLQ4 A0A3Q3CKF4 A0A3B4X271 A0A2U9BJ26 A0A3P8VLC7 A0A3Q4MPV5 A0A1A7XCB5 A0A1A7YGG4 A0A1A8J218 A0A1A8UL30 A0A1W0WL65 A0A151WL97 H0YPC0 A0A093FLT7 A0A3P9HNX1 A0A3Q2TSG4 A0A3L8RUM3 I3JTA5 A0A091G9X6 A0A087QJJ0 A0A3Q2TWE1 U3JH43 A0A091FID0 A0A218UGY0 A0A091RQJ4 R0JIJ9 A0A3Q1GXA1 A0A3M0J4J1 H2MS81
PDB
5VHD
E-value=2.56381e-80,
Score=763
Ontologies
GO
PANTHER
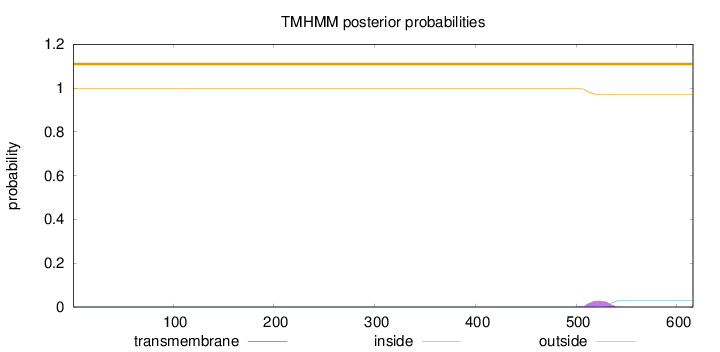
Topology
Length:
616
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.64863
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00046
outside
1 - 616
Population Genetic Test Statistics
Pi
190.900786
Theta
146.344313
Tajima's D
0.764471
CLR
0.19825
CSRT
0.585870706464677
Interpretation
Uncertain
