Gene
KWMTBOMO16398
Pre Gene Modal
BGIBMGA013848
Annotation
serine_protease_inhibitor_32_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.36 PlasmaMembrane Reliability : 1.575
Sequence
CDS
ATGTGGAAGATTGCGTTGACCTTGGCGGTCGTCGTTACATCCGCGCCTCTCGACAATGCGGACTATTCGTCTATAAACACATTCGCATTGCGACTATTGGATCACACTTATGCGTTTCAAGAAGCATTTGGCGTTAAAAACATTGCAATATCACCCCTATCGATTTGGAGCGTGTTCTCGTTGCTGGCGGAGGGTTCGTCTGGAGACACGTTCATGGAATTGCTAAACGCTTTGCAATTACCGAACGATTTAGCCGCGACGCAAACGCTCCATCTGGCCGCCCGCGGTATCTTCAAGTCTACGTCCAGAGACCTTATCCTGAAGGGTCAATCGGTCATGCTCTCCGATTGCTCATTGGAGATCCACCCGGAGTTCTGTGAGCTCGCTAATCTGTACAGCACGGATATATACACGGTCGACCCGAGAAACACGACCAAACTTGCGGCCGATATAAATTATTGGATATGCTTGGCGACAGAAGGAAGAATACTGAATTCGGTTAAACCGCAAGATTTGGAGAACTTAAGGATGGTCCTTGTTGATGCGTTGTATTTTAAAGCGAATTGGAAGTATCCTTTCGATCCGACCCAGACGAAGGAAGAAGCGTTTTATAATTCCCAAGGACAAGTCGTTGGTTCTGTCAATATGATGTACCACAAAGCACCGCACAACCTAGCGGATTCCCCACGAATCGGCGCTCAAGTCTTAGAAATGACTTACGGAAAAGACGAAGAATTTTCAATGCTGATCCTTGTGCCTTTCGACGGAATGCCAATCAAGAAGCTGCTAAACAACTTGGCAACACAGCCGATGGATTGGCTCGAAGACTTCAGACTTGATGACCCCCCAAAAGTGGACTGCTACGTTCCACGTTTTAAAATATCATCGCAATCGGACATGGTAACTCCTCTAAAATACACAGGGATACATTCTATATTCGATGGGCAGAAAGCCGAGTTACCGGGGGTGTCTACAAGTCCGCTCTTTGTGTCATCGGCGATCCAGAACGTAGATTTGGAAGTTACAGAAGAGGGTACCGTGGCGGCTGTGGCCACCGTGGTCGGATTAGAAGACAGGATATTAGGGCCCAGGATCGAGGTGAATAAAGAGTTCGTCTTTCTGATCGTGGACAGAAGGACAAACTTAATAGTGTTCTCTGGCGTCTACGGCGACCCTACAACTGTTTAG
Protein
MWKIALTLAVVVTSAPLDNADYSSINTFALRLLDHTYAFQEAFGVKNIAISPLSIWSVFSLLAEGSSGDTFMELLNALQLPNDLAATQTLHLAARGIFKSTSRDLILKGQSVMLSDCSLEIHPEFCELANLYSTDIYTVDPRNTTKLAADINYWICLATEGRILNSVKPQDLENLRMVLVDALYFKANWKYPFDPTQTKEEAFYNSQGQVVGSVNMMYHKAPHNLADSPRIGAQVLEMTYGKDEEFSMLILVPFDGMPIKKLLNNLATQPMDWLEDFRLDDPPKVDCYVPRFKISSQSDMVTPLKYTGIHSIFDGQKAELPGVSTSPLFVSSAIQNVDLEVTEEGTVAAVATVVGLEDRILGPRIEVNKEFVFLIVDRRTNLIVFSGVYGDPTTV
Summary
Similarity
Belongs to the serpin family.
Uniprot
C0J8I1
A0A0L7LSG9
E1B2D5
G9F9J7
A0A2H1W568
A0A2A4K719
+ More
A0A0N1ID49 A0A0N1PGT8 A0A212EV22 A0A0L7LHE9 A0A2H1W4N9 D2KQN9 Q6Q2D6 R4TUT4 A0A0M4JPK9 Q6Q2D7 A0A1E1WH84 Q6Q2D8 A0A0N1IMR0 A0A2A4K5E1 S4PH08 A0A2W1BCV0 A0A0N1PJL4 A0A1L8DP46 A0A1L8DPF0 A0A1L8DPG3 A0A1L8DPH6 A0A1Y1KYZ6 A0A0L7LTV5 I4DMG6 I4DN70 N6TYC6 A0A0N1PFL7 A0A0M5KHD7 J7HF01 Q6Q2D5 A0A0L7KUV3 A0A1V1FQE8 A0A1B0CTS9 A0A2A4K6S3 A0A1E1WPC2 W8BHP8 A0A2J7QUH4 A0A2J7QUH8 A0A212F0H6 A0A0T6AXD6 A0A2H1VIL1 A0A290U611 W5JE82 Q6Q2D3 I4DN94 T1PGK0 A0A0L7LHI6 A0A2H1W4Y7 A0A0N1I9J2 A0A1L5J018 I4DQD9 A0A1J1IFF1 A0A2W1B5Y5 A0A1I8MAV0 A0A0K8U8K9 A0A1Q3FI97 A0A0K8UPF1 A0A067R1I1 U5EXE5 A0A1L8DPI9 B0X107 A0A3G1VCH4 A0A0T6AXN2 A0A182J5L1 A0A034VN94 A0A0N0PEL1 A0A182K012 A0A182XE44 Q005M7 A0A182HJ80 A0A182RC49 A0A1W4WKL2 A0A182VGH0 Q2PQP4 J7HL87 A0A2A4K8U7 A0A182PT21 A0A182QX10 A0A1B0FBM1 A0A0L0CDH7 A0A1B0ARB3 A0A182U6B2 A0A1A9ZFX6 Q7QBF4 A0A182LS71 H9JWD5 F5HL11 A0A290U612 D6W777 A0A2W1BIT9 H9TZT8 A0A182FKU8 A0A0L7K494 B4MLK8
A0A0N1ID49 A0A0N1PGT8 A0A212EV22 A0A0L7LHE9 A0A2H1W4N9 D2KQN9 Q6Q2D6 R4TUT4 A0A0M4JPK9 Q6Q2D7 A0A1E1WH84 Q6Q2D8 A0A0N1IMR0 A0A2A4K5E1 S4PH08 A0A2W1BCV0 A0A0N1PJL4 A0A1L8DP46 A0A1L8DPF0 A0A1L8DPG3 A0A1L8DPH6 A0A1Y1KYZ6 A0A0L7LTV5 I4DMG6 I4DN70 N6TYC6 A0A0N1PFL7 A0A0M5KHD7 J7HF01 Q6Q2D5 A0A0L7KUV3 A0A1V1FQE8 A0A1B0CTS9 A0A2A4K6S3 A0A1E1WPC2 W8BHP8 A0A2J7QUH4 A0A2J7QUH8 A0A212F0H6 A0A0T6AXD6 A0A2H1VIL1 A0A290U611 W5JE82 Q6Q2D3 I4DN94 T1PGK0 A0A0L7LHI6 A0A2H1W4Y7 A0A0N1I9J2 A0A1L5J018 I4DQD9 A0A1J1IFF1 A0A2W1B5Y5 A0A1I8MAV0 A0A0K8U8K9 A0A1Q3FI97 A0A0K8UPF1 A0A067R1I1 U5EXE5 A0A1L8DPI9 B0X107 A0A3G1VCH4 A0A0T6AXN2 A0A182J5L1 A0A034VN94 A0A0N0PEL1 A0A182K012 A0A182XE44 Q005M7 A0A182HJ80 A0A182RC49 A0A1W4WKL2 A0A182VGH0 Q2PQP4 J7HL87 A0A2A4K8U7 A0A182PT21 A0A182QX10 A0A1B0FBM1 A0A0L0CDH7 A0A1B0ARB3 A0A182U6B2 A0A1A9ZFX6 Q7QBF4 A0A182LS71 H9JWD5 F5HL11 A0A290U612 D6W777 A0A2W1BIT9 H9TZT8 A0A182FKU8 A0A0L7K494 B4MLK8
Pubmed
EMBL
BABH01029197
EU935633
ACG61195.1
JTDY01000179
KOB78425.1
HQ149330
+ More
ADM86478.1 JN033742 AEW46895.1 ODYU01006385 SOQ48188.1 NWSH01000108 PCG79462.1 KQ459680 KPJ20463.1 KQ459547 KPJ00031.1 AGBW02012254 OWR45342.1 JTDY01001104 KOB74872.1 ODYU01006002 SOQ47464.1 GU270470 ACZ81437.1 BABH01029205 AY566164 AAS68505.1 KC686693 AGM20455.1 KR003727 ALD62505.1 AY566163 AAS68504.1 GDQN01010706 GDQN01004833 JAT80348.1 JAT86221.1 AY566162 AAS68503.1 KPJ00036.1 PCG79465.1 GAIX01003432 JAA89128.1 KZ150265 PZC71705.1 KPJ20465.1 GFDF01005831 JAV08253.1 GFDF01005830 JAV08254.1 GFDF01005817 JAV08267.1 GFDF01005829 JAV08255.1 GEZM01069147 JAV66623.1 JTDY01000095 KOB78895.1 AK402484 BAM19106.1 AK402746 BAM19360.1 APGK01047281 KB741077 KB632432 ENN74325.1 ERL95537.1 KPJ00034.1 KR003728 ALD62506.1 JN400443 AFQ01142.1 AY566165 AAS68506.1 JTDY01005636 KOB66799.1 FX985330 BAX07343.1 AJWK01027949 PCG79463.1 GDQN01002297 JAT88757.1 GAMC01005750 JAC00806.1 NEVH01010582 PNF32233.1 PNF32236.1 AGBW02011097 OWR47217.1 LJIG01022579 KRT79823.1 ODYU01002563 SOQ40242.1 KY680238 ATD13319.1 ADMH02001488 ETN62366.1 AY566166 AY566167 AAS68507.1 AAS68508.1 AK402782 BAM19384.1 KA646983 AFP61612.1 KOB74875.1 ODYU01006359 SOQ48141.1 KPJ00033.1 KX117918 APM86796.1 AK404064 BAM20129.1 CVRI01000048 CRK98999.1 KZ150289 PZC71592.1 GDHF01029312 JAI23002.1 GFDL01007765 JAV27280.1 GDHF01025876 GDHF01023735 GDHF01008477 JAI26438.1 JAI28579.1 JAI43837.1 KK852777 KDR16770.1 GANO01000993 JAB58878.1 GFDF01005834 JAV08250.1 DS232250 EDS38399.1 MG732912 AYK02794.1 KRT79825.1 GAKP01015919 JAC43033.1 KQ459764 KPJ20096.1 DQ974167 ABJ52807.1 APCN01002129 DQ307179 ABC25079.1 JN400444 AFQ01143.1 NWSH01000031 PCG80499.1 AXCN02000179 CCAG010010279 JRES01000521 KNC30448.1 JXJN01002301 AAAB01008879 EAA08472.4 AXCM01004089 BABH01029200 EGK96972.1 KY680239 ATD13320.1 KQ971307 EFA11609.1 PZC71593.1 JQ312069 AFG28186.1 JTDY01010789 KOB58011.1 CH963847 EDW72864.2
ADM86478.1 JN033742 AEW46895.1 ODYU01006385 SOQ48188.1 NWSH01000108 PCG79462.1 KQ459680 KPJ20463.1 KQ459547 KPJ00031.1 AGBW02012254 OWR45342.1 JTDY01001104 KOB74872.1 ODYU01006002 SOQ47464.1 GU270470 ACZ81437.1 BABH01029205 AY566164 AAS68505.1 KC686693 AGM20455.1 KR003727 ALD62505.1 AY566163 AAS68504.1 GDQN01010706 GDQN01004833 JAT80348.1 JAT86221.1 AY566162 AAS68503.1 KPJ00036.1 PCG79465.1 GAIX01003432 JAA89128.1 KZ150265 PZC71705.1 KPJ20465.1 GFDF01005831 JAV08253.1 GFDF01005830 JAV08254.1 GFDF01005817 JAV08267.1 GFDF01005829 JAV08255.1 GEZM01069147 JAV66623.1 JTDY01000095 KOB78895.1 AK402484 BAM19106.1 AK402746 BAM19360.1 APGK01047281 KB741077 KB632432 ENN74325.1 ERL95537.1 KPJ00034.1 KR003728 ALD62506.1 JN400443 AFQ01142.1 AY566165 AAS68506.1 JTDY01005636 KOB66799.1 FX985330 BAX07343.1 AJWK01027949 PCG79463.1 GDQN01002297 JAT88757.1 GAMC01005750 JAC00806.1 NEVH01010582 PNF32233.1 PNF32236.1 AGBW02011097 OWR47217.1 LJIG01022579 KRT79823.1 ODYU01002563 SOQ40242.1 KY680238 ATD13319.1 ADMH02001488 ETN62366.1 AY566166 AY566167 AAS68507.1 AAS68508.1 AK402782 BAM19384.1 KA646983 AFP61612.1 KOB74875.1 ODYU01006359 SOQ48141.1 KPJ00033.1 KX117918 APM86796.1 AK404064 BAM20129.1 CVRI01000048 CRK98999.1 KZ150289 PZC71592.1 GDHF01029312 JAI23002.1 GFDL01007765 JAV27280.1 GDHF01025876 GDHF01023735 GDHF01008477 JAI26438.1 JAI28579.1 JAI43837.1 KK852777 KDR16770.1 GANO01000993 JAB58878.1 GFDF01005834 JAV08250.1 DS232250 EDS38399.1 MG732912 AYK02794.1 KRT79825.1 GAKP01015919 JAC43033.1 KQ459764 KPJ20096.1 DQ974167 ABJ52807.1 APCN01002129 DQ307179 ABC25079.1 JN400444 AFQ01143.1 NWSH01000031 PCG80499.1 AXCN02000179 CCAG010010279 JRES01000521 KNC30448.1 JXJN01002301 AAAB01008879 EAA08472.4 AXCM01004089 BABH01029200 EGK96972.1 KY680239 ATD13320.1 KQ971307 EFA11609.1 PZC71593.1 JQ312069 AFG28186.1 JTDY01010789 KOB58011.1 CH963847 EDW72864.2
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000019118 UP000030742 UP000092461 UP000235965 UP000000673 UP000183832 UP000095301 UP000027135 UP000002320 UP000075880 UP000075881 UP000076407 UP000075840 UP000075900 UP000192223 UP000075903 UP000075885 UP000075886 UP000092444 UP000037069 UP000092460 UP000075902 UP000092445 UP000007062 UP000075883 UP000007266 UP000069272 UP000007798
UP000019118 UP000030742 UP000092461 UP000235965 UP000000673 UP000183832 UP000095301 UP000027135 UP000002320 UP000075880 UP000075881 UP000076407 UP000075840 UP000075900 UP000192223 UP000075903 UP000075885 UP000075886 UP000092444 UP000037069 UP000092460 UP000075902 UP000092445 UP000007062 UP000075883 UP000007266 UP000069272 UP000007798
Pfam
PF00079 Serpin
Interpro
SUPFAM
SSF56574
SSF56574
Gene 3D
ProteinModelPortal
C0J8I1
A0A0L7LSG9
E1B2D5
G9F9J7
A0A2H1W568
A0A2A4K719
+ More
A0A0N1ID49 A0A0N1PGT8 A0A212EV22 A0A0L7LHE9 A0A2H1W4N9 D2KQN9 Q6Q2D6 R4TUT4 A0A0M4JPK9 Q6Q2D7 A0A1E1WH84 Q6Q2D8 A0A0N1IMR0 A0A2A4K5E1 S4PH08 A0A2W1BCV0 A0A0N1PJL4 A0A1L8DP46 A0A1L8DPF0 A0A1L8DPG3 A0A1L8DPH6 A0A1Y1KYZ6 A0A0L7LTV5 I4DMG6 I4DN70 N6TYC6 A0A0N1PFL7 A0A0M5KHD7 J7HF01 Q6Q2D5 A0A0L7KUV3 A0A1V1FQE8 A0A1B0CTS9 A0A2A4K6S3 A0A1E1WPC2 W8BHP8 A0A2J7QUH4 A0A2J7QUH8 A0A212F0H6 A0A0T6AXD6 A0A2H1VIL1 A0A290U611 W5JE82 Q6Q2D3 I4DN94 T1PGK0 A0A0L7LHI6 A0A2H1W4Y7 A0A0N1I9J2 A0A1L5J018 I4DQD9 A0A1J1IFF1 A0A2W1B5Y5 A0A1I8MAV0 A0A0K8U8K9 A0A1Q3FI97 A0A0K8UPF1 A0A067R1I1 U5EXE5 A0A1L8DPI9 B0X107 A0A3G1VCH4 A0A0T6AXN2 A0A182J5L1 A0A034VN94 A0A0N0PEL1 A0A182K012 A0A182XE44 Q005M7 A0A182HJ80 A0A182RC49 A0A1W4WKL2 A0A182VGH0 Q2PQP4 J7HL87 A0A2A4K8U7 A0A182PT21 A0A182QX10 A0A1B0FBM1 A0A0L0CDH7 A0A1B0ARB3 A0A182U6B2 A0A1A9ZFX6 Q7QBF4 A0A182LS71 H9JWD5 F5HL11 A0A290U612 D6W777 A0A2W1BIT9 H9TZT8 A0A182FKU8 A0A0L7K494 B4MLK8
A0A0N1ID49 A0A0N1PGT8 A0A212EV22 A0A0L7LHE9 A0A2H1W4N9 D2KQN9 Q6Q2D6 R4TUT4 A0A0M4JPK9 Q6Q2D7 A0A1E1WH84 Q6Q2D8 A0A0N1IMR0 A0A2A4K5E1 S4PH08 A0A2W1BCV0 A0A0N1PJL4 A0A1L8DP46 A0A1L8DPF0 A0A1L8DPG3 A0A1L8DPH6 A0A1Y1KYZ6 A0A0L7LTV5 I4DMG6 I4DN70 N6TYC6 A0A0N1PFL7 A0A0M5KHD7 J7HF01 Q6Q2D5 A0A0L7KUV3 A0A1V1FQE8 A0A1B0CTS9 A0A2A4K6S3 A0A1E1WPC2 W8BHP8 A0A2J7QUH4 A0A2J7QUH8 A0A212F0H6 A0A0T6AXD6 A0A2H1VIL1 A0A290U611 W5JE82 Q6Q2D3 I4DN94 T1PGK0 A0A0L7LHI6 A0A2H1W4Y7 A0A0N1I9J2 A0A1L5J018 I4DQD9 A0A1J1IFF1 A0A2W1B5Y5 A0A1I8MAV0 A0A0K8U8K9 A0A1Q3FI97 A0A0K8UPF1 A0A067R1I1 U5EXE5 A0A1L8DPI9 B0X107 A0A3G1VCH4 A0A0T6AXN2 A0A182J5L1 A0A034VN94 A0A0N0PEL1 A0A182K012 A0A182XE44 Q005M7 A0A182HJ80 A0A182RC49 A0A1W4WKL2 A0A182VGH0 Q2PQP4 J7HL87 A0A2A4K8U7 A0A182PT21 A0A182QX10 A0A1B0FBM1 A0A0L0CDH7 A0A1B0ARB3 A0A182U6B2 A0A1A9ZFX6 Q7QBF4 A0A182LS71 H9JWD5 F5HL11 A0A290U612 D6W777 A0A2W1BIT9 H9TZT8 A0A182FKU8 A0A0L7K494 B4MLK8
PDB
6EE5
E-value=2.40563e-29,
Score=321
Ontologies
PANTHER
Topology
SignalP
Position: 1 - 14,
Likelihood: 0.952939
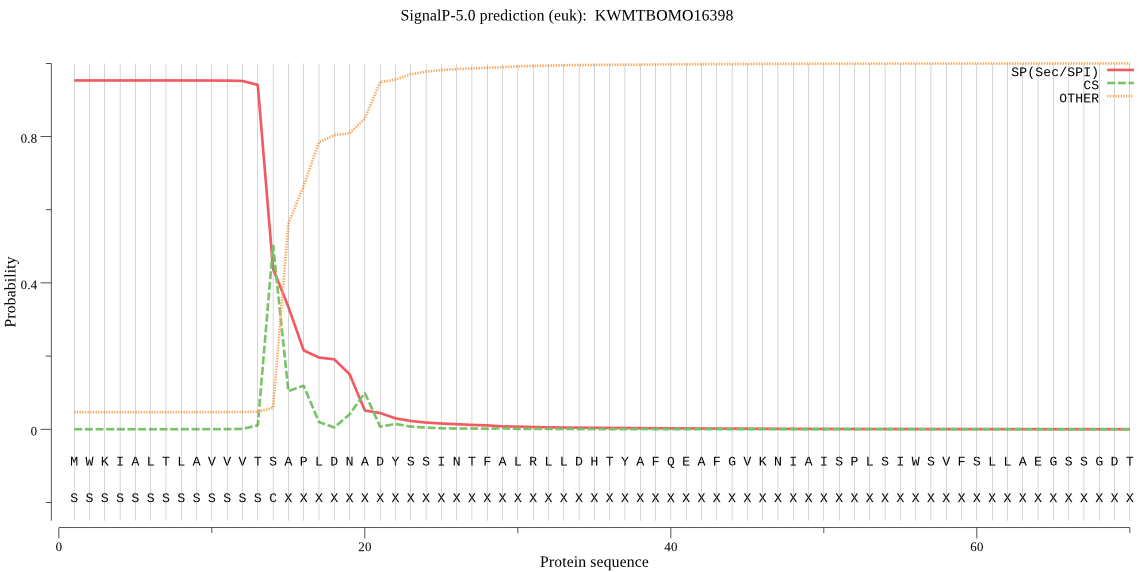
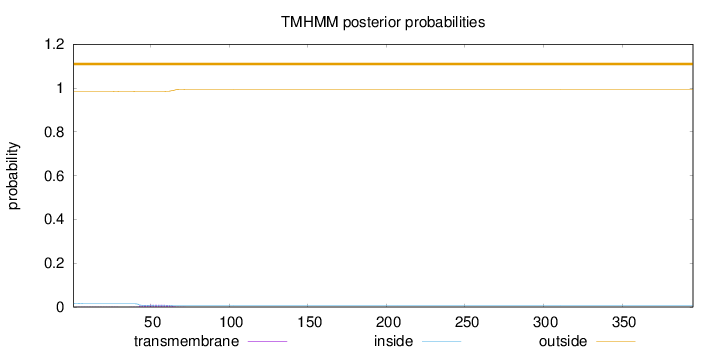
Length:
395
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.22163
Exp number, first 60 AAs:
0.18208
Total prob of N-in:
0.01565
outside
1 - 395
Population Genetic Test Statistics
Pi
253.62233
Theta
208.638371
Tajima's D
0.63414
CLR
0.163064
CSRT
0.552822358882056
Interpretation
Uncertain
