Pre Gene Modal
BGIBMGA013841
Annotation
PREDICTED:_autophagy-related_protein_13_homolog_[Papilio_polytes]
Full name
Autophagy-related protein 13
Location in the cell
Nuclear Reliability : 2.882
Sequence
CDS
ATGGACAAAAGTGCTACATTACAAGAGCTCGGCTACAAAGACGAGAATGAGTTGAGAGAGACAATATATGACTATTTGAGGACAATTCGAGACCCCGAAAAGCCAAATACGCTTGAAGACTTGAAGGTCGTGTACGAGGAAGGAATCTTTGTGAAGGAACCAACTGCAGATAAAGTGCCCGTTTTACGTGTGGAGTATAATCCAACAGTGCCCCATTGTTCGCTGGCCACACTAATCGGACTTTGCATTCGTATTAAAATATTAAGGTCTATCCATCATCCTGTGAAATTGGACATTTTTATCAAGAAAGGGGCACATACCACTGAAGATGAAATCAATAAGCAGATCAACGACAAGGAGAGGATTGCAGCTGCAATGGAGAATCCCAATCTAAGAAACCTGGTGGAAAACTGCATAGCCGAAGAGGATAAACCAAAGACGTCAGACACTGATTGGTTTAATTTACAGATACCAGATTCCTCTGAAGTAAATCAGGCAACTAAGAATGCTTTACCTTCGGATCGTGTACTGGAGACGATAAAATCACAATTACATGTAGAAATATCTGTTCAAACAGAAGATGGAGATGAAATGGTACTAGAACTCTGGACTCTTCAGCTAGACGAAACCCAATTCGATACATCATTGAAGGCAATGAACACAGTGTATTTTAGAATGGGCATATTGTTAAAATCTCTTATAACTATAACAAGGATCACACCGGCATATCATTTGTCAAGGAAACAGAGACTGGAATCATTCACTATATTCTACAGAGTTTACAGTGGCGAGGCAAAATTGAAAGCTCTTGGTGATTCAGTGAAAACTATTCAAGCCGGCCTCTTAAAAACACCATTAGGTGCTTTAGTCTTCTCGGTAACGTACCGTACAAATTTTTCAATAACTCCTAACCGTTCAGACAATAACAAAGCATTGTTATTAAAGAGTGATCACTTTGAATTGAGTCCGAAACATGTCATATTTGAATCAAAAAAGAAGAAAGAAGAAAAAAGAGAGTCGAAACCTTCAGGACCCATCGATTTGGACAAGCCCTTACGCTTAGCTGCTTTTGTAGATGAAACCTTAGTTAAGAGGACATTTGAAGAATTCTACACGAAAATTCCTATTCCTGATCACTACAGACCTGAAAAATTGCCTGTATCTCCTGTTAAAGAAAAGGAAATCATGTCTAAGAGCACACAAGAATTAGATGACAGTGCAGCATCAAAGAGTCCGAATTCTTTAGAGATGCCACCAAAGAAATTCTCTGGATTCCGTCACGAAAACGAACCACCAATAAAACTGTTATTGTTCCCTTTTGCTGACACACATCCTATACGAGAACTAGCCGAGTTTTACAAAGATTTCTTTAATGCGCCACACTTGAAGTTGACAGATGAGCTAACACAATCGAACAAAAGCTTAGAGCCTGTTTTAGAAGACGTAGAACTAACCAACGAAGATTTGTCTAAAGATTTGGAGCTGTATGAGAATTCAGTTGTTGAGTTTGATGAATTGTTGGCCGACATGTGTAGATCGGCTGAGTGGAGTGGGAATTAA
Protein
MDKSATLQELGYKDENELRETIYDYLRTIRDPEKPNTLEDLKVVYEEGIFVKEPTADKVPVLRVEYNPTVPHCSLATLIGLCIRIKILRSIHHPVKLDIFIKKGAHTTEDEINKQINDKERIAAAMENPNLRNLVENCIAEEDKPKTSDTDWFNLQIPDSSEVNQATKNALPSDRVLETIKSQLHVEISVQTEDGDEMVLELWTLQLDETQFDTSLKAMNTVYFRMGILLKSLITITRITPAYHLSRKQRLESFTIFYRVYSGEAKLKALGDSVKTIQAGLLKTPLGALVFSVTYRTNFSITPNRSDNNKALLLKSDHFELSPKHVIFESKKKKEEKRESKPSGPIDLDKPLRLAAFVDETLVKRTFEEFYTKIPIPDHYRPEKLPVSPVKEKEIMSKSTQELDDSAASKSPNSLEMPPKKFSGFRHENEPPIKLLLFPFADTHPIRELAEFYKDFFNAPHLKLTDELTQSNKSLEPVLEDVELTNEDLSKDLELYENSVVEFDELLADMCRSAEWSGN
Summary
Similarity
Belongs to the ATG13 family. Metazoan subfamily.
Uniprot
EMBL
JTDY01009683
KOB63047.1
NWSH01000450
PCG76328.1
KQ459054
KPJ03922.1
+ More
KQ460047 KPJ17991.1 GDQN01003364 GDQN01003281 GDQN01003118 GDQN01000958 JAT87690.1 JAT87773.1 JAT87936.1 JAT90096.1 ODYU01006546 SOQ48485.1 CVRI01000063 CRL04695.1 GFDF01003760 JAV10324.1 GFDF01003759 JAV10325.1 GANO01001632 JAB58239.1 PYGN01000819 PSN40375.1
KQ460047 KPJ17991.1 GDQN01003364 GDQN01003281 GDQN01003118 GDQN01000958 JAT87690.1 JAT87773.1 JAT87936.1 JAT90096.1 ODYU01006546 SOQ48485.1 CVRI01000063 CRL04695.1 GFDF01003760 JAV10324.1 GFDF01003759 JAV10325.1 GANO01001632 JAB58239.1 PYGN01000819 PSN40375.1
Proteomes
Interpro
SUPFAM
SSF117916
SSF117916
Gene 3D
ProteinModelPortal
PDB
2M5H
E-value=3.21245e-34,
Score=364
Ontologies
PATHWAY
GO
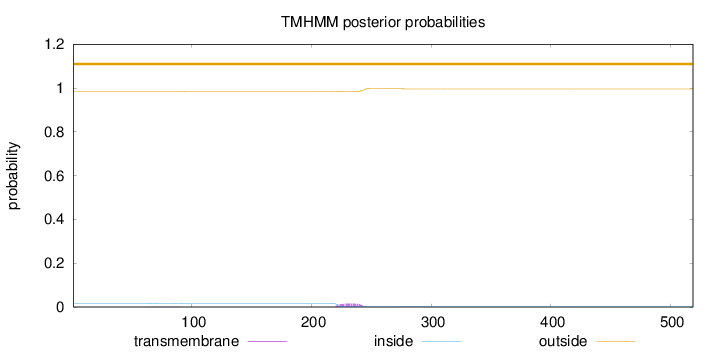
Topology
Length:
519
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.35507
Exp number, first 60 AAs:
5e-05
Total prob of N-in:
0.01549
outside
1 - 519
Population Genetic Test Statistics
Pi
185.665227
Theta
210.911894
Tajima's D
-0.578398
CLR
1.006867
CSRT
0.227888605569722
Interpretation
Uncertain
