Pre Gene Modal
BGIBMGA013842
Annotation
PREDICTED:_cleavage_and_polyadenylation_specificity_factor_subunit_1_[Bombyx_mori]
Full name
Cleavage and polyadenylation specificity factor subunit 1
Alternative Name
Cleavage and polyadenylation specificity factor 160 kDa subunit
Location in the cell
PlasmaMembrane Reliability : 2.269
Sequence
CDS
ATGTTTTCGATTTGTCGAACGACTCATCCAGCGACTGGTATAGAGCACGCGGTTAGCTGTTATTTCTTTAACAATGACGAGGTTTGTCTGGTCACCGCCGGTGCCAATATCATCAAGGTCTTCAGGCTGCTGCCCGAAGGCCACACAAAAGAGATTAATGCAGCCGGTCAGCCGATTCCCCCGAAGATGAAGTTGGAATGCTTAGCGACATATACATTATGGGGTAATGTGATGTCTTTAGCTTCTGTACGCTGTCAAAGTTCTGGTCGAGATATCCTGTTGGTTTCATTCAAAGAAGCTAAACTATCTGTGCTCCAATATGATCCACAGACCAATAACTTGGTGACATTGAGTATGCATTATTTTGAAGAGGATGATATGAAGGGCGGTTGGACAACTCATCCTCATATACCATGGATCAGAGTAGATCCTGAGTTCCGATGTGCTGTGATGTTGTTGTATGGGAAGAAACTTGCAGTCCTTCCGTTTAGGAAGGACATTACCTCTGAAGAAGGGGATCCTCTTGAGGCCAAACCCCTCTCTGACACAAAGAAAAATCAAGCAGCTCATACTATGAACAGGGCACCAACACTGGCCTCATACATAATAATACTCAAGGACCTAGACGAGAAGATTGATAACATATTAGACATACAATTCTTGCATGGCTATTACGAGCCTACTTTGCTACTGCTCTATGAGCCTGTCAGAACATTTCCTGGTCGCACGGCGGTTCGCAACGACACCTGCGCGATGGCGGGCGTCAGTCTGAACATGTCAGCTCGGGTGCACCCGGTCATCTGGTCCACCTCAGGTCTGCCCTTCGACTGTCTCCAAGCCGTGCCCGTACCGAAACCACTAGGTGGCTGTTTGATAATGGCCGTCAACTCATTGATATATTTAAATCAATCGGTTCCACCTTACGGCGTCTCACTGAACAGTGTTGCCACACACACCACAAATTTTCCCCTGAGGATCCAGGAGGGCGTGTGCATGACGCTGGAGGGCGCGTGCGTGGCGCGGCTGTCGGACGCCGTGTTCGCGGTGTCTCTGCGCGGCGGCCAGCTGTACGTTGTGGCGCTGCTGGCGGACAGTGTGCGCTCCGTCAGAAGCTTCCACATCGACCGCGCCGCCGCCTCCGTGCTCACCACCTGCATGTGCGTTATAGAAGAGGACTTCCTTTTCCTGGGGTCGCGGCTCGGAAACTCGCTGCTGCTGCGAGTTACCGAGCGAGAGAACCGGATGCTGTTCTCCGTTGACAAGCCCCTGGAAGCGACGGTCGATCTGACCGTGGCCGAAACCGAACGAGAGCAGAGCAGAGAGAAGGAGCAGCCCGACCCGGCCTCGAAGCGGCGGCGCATGGACACCATCAGCGACTGCGTGGCCTCCAACGTCATCGAGATCAGCGACGGAGACGAGCTCGAGGTGTACGGCAGCGACATACGCACCTCCACAAGACTCACCAGCTACGTGTTCGAGGTATGCGATTCGCTGTTAAACATATGTCCGATCGGCGACGTGTCTATGGGAGAGCCCCAGCTCCTCTCCGAGGAGTACAGCAGGCAGCAGGAGAATCCTCTGGTAGAACTGGTGACCTGTAGTGGCAGGGGGAAGAACGGCGCCCTGACCGTGCTGCAGAGGACGGTCAGACCTCAGCTGATTACTGCTTTCAATTTACCCGGCTGTATCGACATGTGGTCGGTTTTCGGAGAGAGCGAAAGCACCAGAGAAAACGAAGGGACGCACGCGTATTTGATACTGACGCAGGACGACTCCTCCATGGTTCTGCAAACTGGCCAAGAGATCAACGAGGTGGACAACTCTGGCTTCATGACGGGTTCGCCCACCATCTTCGCCGGAAACCTCGGGAATAACAAATTCATGGTGCAGGTCACCACCACCACCATTCGACTCATCAAGGGCGGCGTGCAGCTGCAGTCGATCCAGCTGGAGTGGACGGCGCAGTACGCGGCCGTCGCGGACCCCTACCTGTGCGTCACGTCCAGCTGCGGCCGCGCCATCGTGCTGGCCCTGAGAGAGATCAGGGGGAAGGACGGGCAGCCGTCGGGTCGGCTGGCGCCCACGCGGCAGTGCGTCCCCGCGCGGCCCCCGCTCGTGCGCGCCGCCCCGCACCGCGACCTCAGCGGACTGTTCGCGCCAGACCTCGCCGAGCCCGGCCACCAGGTCAAAGGTGAGTTCACCGGTAAGCTGAAGGAGAACGTGAAGAAGGAGGCCTTCAAGCCCGGCGTGGTCTATGAGCTGAACGACGAAGACGAGATGCTGTACGGAGGCGACCAGACACCTGCATCCATGGCCAGCATAATGCTGGCGGAGCAAAGCAAGAATCCAGGGGTTCCGCGTCGGATGTCGCGCTGGTGGAAGAAGTACCTGCAGGAGGTGAAGCCCACGTACTGGCTGTTCTGCGTTCGCGACAACGGCAACCTGGAGATATACTCGTTGCCGGAAATGAGGTTGAGTTTCCTGGTGAGAGACTCGTGCGCTGGAAACAGGATATTGGCGGACAGCCTGGAGTCCGTCCCGATGTCAGACAGCATGGAGGACGAGGACATCAGCGCGCCGTCACACAACGCTGATGCTGAGAAGCTCAAGGAACTGTGCGTCGTCGGTCTGGGGCACAAGGGTTCCAGGGTGATACTGATGCTGAGGATGCAAAATGAATTGATGATTTATCAGGCGTACAAATACCCCCGGGGGAATCTCAAGCTGCGGTTCTGCCGGGTGACTGTCTCGTTCCCGTTTGGGTACGAGAGCGTGCCCAGCACCGCACAGAGCGCCGACACCTACGAGACGGCCGGCGTGCGGGACGGGGTGCGGCAGCTGCGGTACTTTGGCAACGTGGGCGGGTACAGCGGCGTGTTCGTCTGCGGCGCGACCCCCTACATGATGTTCTTGTCTGCGCGCGGCGAGCTGCGCCTGCACCCGCTGCACGCCGACCGACCGCCCGTGCACACGTTCGCGCCCTTCAACAACACCAATTGTCCGCAGGGCTTCCTGTACTTTAACGCCGAGTCGTCGCTCCGCATCTGCGCGCTGCCGTCGCACCTGTCGTACGACGCGGCGTGGGCGGTGCGCAAGGTGCCCATCCGCATGACGCCGCACTACGTCACGTTCCACCTGGAGTCCAGGACCTACTGCCTCGTGGCCTCCACCTCGCAGCCCACCACGGTCTACTACAAGTTCAACGGCGAGGACAAGGAGAAGTCTGCCGAGAACAAAGGGGACAGGTTCCCGTACCCGATGCTGGATCGCTTCTCGGTGATGCTGTTCTCCCCCGTCTCCTGGGAGATCATACCGAACACCAAGATCGAGCTGGACGAGTGGGAGCACGTGACGTGTCTCAAGAACGTGTCGCTGTCGTACGAAGGAACCAGATCCGGTCTCCGCGGTTACATAGCGATAGGAACTAATTATAATTACAGCGAGGATATCACTTCTAGGGGGCGGATAATAATCTACGACATAATCGACGTGGTCCCGGAGCCCGGGCAGCCGCTGACCAAGAACCGGTTCAAGGAGATCTACTCCAAGGAGCAGAAGGGTCCGGTGACCGCGCTCACGCAGGTGCTCGGCTACCTCATATCCGCCGTCGGACAGAAGATATACATATGGCAGCTGAAGGACAACGACCTAGTGGGCGTCGCGTTCATCGACACCCAGATATACGTGCACCGCATGCTGGCCGTCAAGAACCTCATCCTGGTGGCCGACGTCTACAAGTCCATCTCGCTGCTGCGCTACCAGCCCGCGCACAGGACGCTCTCGCTGGTCTCCAGGGACTTCAGGACCGCGCAGATATACGATATGGAGTTCATGGTGGATAATACGAATCTCGGTTTCCTGGTCAGCGAAGCCGAGGGGAACTTCGCGCTGTTCATGTACCAGCCGCAGGCCAGGGAGAGCTACGGAGGCCAGCGGCTGATACGCAAGAGCGACTACCACCTGGGCCAGCAGGTGCACGCCATGTTCCGCATCAACGTGCGCTCGCTGCCAGACTCCGACAACTCGCACAAGCGACACGTCAGCATGTTCACGACCCTGGACGGCGCCATAGGCTACGTGCTGCCGGTCACGGAGAAGATGTACCGGCGCCTACTGATGTTGCAGAACGTCATGAACAACTACTACTGCCACATAGCCGGCCTCAACCCGAGGGCCTTTCGCACGTACAAGGCGTCGCGGCGCGCGGCGGGGGGCGGGGCGGCGCGGGGCGTGCTGGACGGGGACCTGGTGGCGCTGTACGCCGCCATGCCGCGCGCCGACCAGCACGACATCGCAAAGAAAATCGGTACAAAAGTCGAGGAGATAATGTCGGACTTGTACGAGATAGACAGACTGACCGCGCATTTTTAG
Protein
MFSICRTTHPATGIEHAVSCYFFNNDEVCLVTAGANIIKVFRLLPEGHTKEINAAGQPIPPKMKLECLATYTLWGNVMSLASVRCQSSGRDILLVSFKEAKLSVLQYDPQTNNLVTLSMHYFEEDDMKGGWTTHPHIPWIRVDPEFRCAVMLLYGKKLAVLPFRKDITSEEGDPLEAKPLSDTKKNQAAHTMNRAPTLASYIIILKDLDEKIDNILDIQFLHGYYEPTLLLLYEPVRTFPGRTAVRNDTCAMAGVSLNMSARVHPVIWSTSGLPFDCLQAVPVPKPLGGCLIMAVNSLIYLNQSVPPYGVSLNSVATHTTNFPLRIQEGVCMTLEGACVARLSDAVFAVSLRGGQLYVVALLADSVRSVRSFHIDRAAASVLTTCMCVIEEDFLFLGSRLGNSLLLRVTERENRMLFSVDKPLEATVDLTVAETEREQSREKEQPDPASKRRRMDTISDCVASNVIEISDGDELEVYGSDIRTSTRLTSYVFEVCDSLLNICPIGDVSMGEPQLLSEEYSRQQENPLVELVTCSGRGKNGALTVLQRTVRPQLITAFNLPGCIDMWSVFGESESTRENEGTHAYLILTQDDSSMVLQTGQEINEVDNSGFMTGSPTIFAGNLGNNKFMVQVTTTTIRLIKGGVQLQSIQLEWTAQYAAVADPYLCVTSSCGRAIVLALREIRGKDGQPSGRLAPTRQCVPARPPLVRAAPHRDLSGLFAPDLAEPGHQVKGEFTGKLKENVKKEAFKPGVVYELNDEDEMLYGGDQTPASMASIMLAEQSKNPGVPRRMSRWWKKYLQEVKPTYWLFCVRDNGNLEIYSLPEMRLSFLVRDSCAGNRILADSLESVPMSDSMEDEDISAPSHNADAEKLKELCVVGLGHKGSRVILMLRMQNELMIYQAYKYPRGNLKLRFCRVTVSFPFGYESVPSTAQSADTYETAGVRDGVRQLRYFGNVGGYSGVFVCGATPYMMFLSARGELRLHPLHADRPPVHTFAPFNNTNCPQGFLYFNAESSLRICALPSHLSYDAAWAVRKVPIRMTPHYVTFHLESRTYCLVASTSQPTTVYYKFNGEDKEKSAENKGDRFPYPMLDRFSVMLFSPVSWEIIPNTKIELDEWEHVTCLKNVSLSYEGTRSGLRGYIAIGTNYNYSEDITSRGRIIIYDIIDVVPEPGQPLTKNRFKEIYSKEQKGPVTALTQVLGYLISAVGQKIYIWQLKDNDLVGVAFIDTQIYVHRMLAVKNLILVADVYKSISLLRYQPAHRTLSLVSRDFRTAQIYDMEFMVDNTNLGFLVSEAEGNFALFMYQPQARESYGGQRLIRKSDYHLGQQVHAMFRINVRSLPDSDNSHKRHVSMFTTLDGAIGYVLPVTEKMYRRLLMLQNVMNNYYCHIAGLNPRAFRTYKASRRAAGGGAARGVLDGDLVALYAAMPRADQHDIAKKIGTKVEEIMSDLYEIDRLTAHF
Summary
Description
Component of the cleavage and polyadenylation specificity factor (CPSF) complex that plays a key role in pre-mRNA 3'-end formation, recognizing the AAUAAA signal sequence and interacting with poly(A) polymerase and other factors to bring about cleavage and poly(A) addition. This subunit is involved in the RNA recognition step of the polyadenylation reaction (By similarity).
Subunit
Component of the cleavage and polyadenylation specificity factor (CPSF) complex, composed of at least Clp, Cpsf73, Cpsf100 and Cpsf160.
Component of the cleavage and polyadenylation specificity factor (CPSF) complex, composed of CPSF1, CPSF2, CPSF3, CPSF4 and FIP1L1. Found in a complex with CPSF1, FIP1L1 and PAPOLA. Interacts with FIP1L1 and SRRM1. Interacts with TUT1; the interaction is direct and mediates the recruitment of the CPSF complex on the 3'UTR of selected pre-mRNAs (By similarity). Interacts with TENT2/GLD2.
Component of the cleavage and polyadenylation specificity factor (CPSF) complex, composed of CPSF1, CPSF2, CPSF3, CPSF4 and FIP1L1. Found in a complex with CPSF1, FIP1L1 and PAPOLA. Interacts with FIP1L1, TENT2/GLD2 and SRRM1. Interacts with TUT1; the interaction is direct and mediates the recruitment of the CPSF complex on the 3'UTR of selected pre-mRNAs (By similarity).
Component of the cleavage and polyadenylation specificity factor (CPSF) complex, composed of CPSF1, CPSF2, CPSF3, CPSF4 and FIP1L1. Found in a complex with CPSF1, FIP1L1 and PAPOLA. Interacts with FIP1L1 and SRRM1. Interacts with TUT1; the interaction is direct and mediates the recruitment of the CPSF complex on the 3'UTR of selected pre-mRNAs (By similarity). Interacts with TENT2/GLD2.
Component of the cleavage and polyadenylation specificity factor (CPSF) complex, composed of CPSF1, CPSF2, CPSF3, CPSF4 and FIP1L1. Found in a complex with CPSF1, FIP1L1 and PAPOLA. Interacts with FIP1L1, TENT2/GLD2 and SRRM1. Interacts with TUT1; the interaction is direct and mediates the recruitment of the CPSF complex on the 3'UTR of selected pre-mRNAs (By similarity).
Similarity
Belongs to the CPSF1 family.
Keywords
Alternative splicing
Complete proteome
mRNA processing
Nucleus
Reference proteome
RNA-binding
Phosphoprotein
Direct protein sequencing
Feature
chain Cleavage and polyadenylation specificity factor subunit 1
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9JWC8
A0A2A4JWZ2
A0A067QMH4
A0A2J7PFU6
A0A2J7PFU9
A0A310SM89
+ More
A0A2A3EAC1 A0A087ZX72 E1ZVR7 A0A158NZY8 A0A3L8DWZ0 K7ITG7 A0A1W4XRH5 D6WFP3 A0A1B6KC09 A0A1B6G369 A0A1Y1L5P4 F4X3Z7 E0VY56 A0A195CQR4 N6TMU2 E9ISL3 A0A195F296 T1HLS2 A0A146M172 A0A154PDP7 A0A232F4C9 A0A151X3V9 A0A151J0U4 A0A224X582 A0A1Q3FWQ1 A0A182Y710 A0A182W1M8 A0A084VHX0 A0A182RLI6 A0A182GK16 Q17N25 A0A182KCV3 Q7QHS8 A0A182INZ6 A0A182LH94 A0A182LV33 A0A1S4H6C5 A0A182P5P9 W5JVI9 A0A182FBZ4 T1J479 A0A182V794 A0A2M4A931 A0A0J7L9D9 A0A182HY45 A0A034VF18 A0A0K8VQE0 E2C9X3 A0A0A1XA51 A0A182WXH0 A0A195BLJ1 A0A1A9W3R0 A0A1I8NUG9 A0A1A9V1N0 A0A1W4U6G1 A0A1J1HKM0 B3ME15 Q291E2 B4GAU6 B4LJU3 A0A0M4E4M5 A0A3B0J601 B4P767 B3NR35 B4KT60 A0A0J9RCL1 B4HRD9 Q9V726 B4NN81 A0A1A9Z1W2 A0A0P4VRN3 A0A131Z3B1 B4QFU9 A0A224YUK0 B4JW88 A0A1B0F9F7 A0A0P6J4F5 A0A0Q9WW52 Q9V726-2 A0A2P2I273 A0A091DK29 A0A1W4XTH9 G5C5S6 H0WG63 A0A1W4Y3B8 A0A1U7QU45 A0A250Y7S1 B5DEL2 A0A0F7ZBG5 Q9EPU4 W5U6H6 A0A0D9R822 Q10569 A0A2J8RHR5
A0A2A3EAC1 A0A087ZX72 E1ZVR7 A0A158NZY8 A0A3L8DWZ0 K7ITG7 A0A1W4XRH5 D6WFP3 A0A1B6KC09 A0A1B6G369 A0A1Y1L5P4 F4X3Z7 E0VY56 A0A195CQR4 N6TMU2 E9ISL3 A0A195F296 T1HLS2 A0A146M172 A0A154PDP7 A0A232F4C9 A0A151X3V9 A0A151J0U4 A0A224X582 A0A1Q3FWQ1 A0A182Y710 A0A182W1M8 A0A084VHX0 A0A182RLI6 A0A182GK16 Q17N25 A0A182KCV3 Q7QHS8 A0A182INZ6 A0A182LH94 A0A182LV33 A0A1S4H6C5 A0A182P5P9 W5JVI9 A0A182FBZ4 T1J479 A0A182V794 A0A2M4A931 A0A0J7L9D9 A0A182HY45 A0A034VF18 A0A0K8VQE0 E2C9X3 A0A0A1XA51 A0A182WXH0 A0A195BLJ1 A0A1A9W3R0 A0A1I8NUG9 A0A1A9V1N0 A0A1W4U6G1 A0A1J1HKM0 B3ME15 Q291E2 B4GAU6 B4LJU3 A0A0M4E4M5 A0A3B0J601 B4P767 B3NR35 B4KT60 A0A0J9RCL1 B4HRD9 Q9V726 B4NN81 A0A1A9Z1W2 A0A0P4VRN3 A0A131Z3B1 B4QFU9 A0A224YUK0 B4JW88 A0A1B0F9F7 A0A0P6J4F5 A0A0Q9WW52 Q9V726-2 A0A2P2I273 A0A091DK29 A0A1W4XTH9 G5C5S6 H0WG63 A0A1W4Y3B8 A0A1U7QU45 A0A250Y7S1 B5DEL2 A0A0F7ZBG5 Q9EPU4 W5U6H6 A0A0D9R822 Q10569 A0A2J8RHR5
Pubmed
19121390
24845553
20798317
21347285
30249741
20075255
+ More
18362917 19820115 28004739 21719571 20566863 23537049 21282665 26823975 28648823 25244985 24438588 26483478 17510324 12364791 20966253 20920257 23761445 25348373 25830018 17994087 15632085 17550304 22936249 9604891 10731132 12537572 12537569 19450530 26830274 28797301 21993625 28087693 15489334 15057822 22673903 11369601 17927953 21183079 23127152 7651824 1756731
18362917 19820115 28004739 21719571 20566863 23537049 21282665 26823975 28648823 25244985 24438588 26483478 17510324 12364791 20966253 20920257 23761445 25348373 25830018 17994087 15632085 17550304 22936249 9604891 10731132 12537572 12537569 19450530 26830274 28797301 21993625 28087693 15489334 15057822 22673903 11369601 17927953 21183079 23127152 7651824 1756731
EMBL
BABH01029195
BABH01029196
NWSH01000450
PCG76329.1
KK853149
KDR10643.1
+ More
NEVH01025648 PNF15214.1 PNF15210.1 KQ761323 OAD57895.1 KZ288309 PBC28675.1 GL434618 EFN74734.1 ADTU01005136 QOIP01000003 RLU24961.1 AAZX01006832 KQ971319 EFA00240.2 GEBQ01030988 JAT08989.1 GECZ01012880 JAS56889.1 GEZM01066160 JAV68121.1 GL888624 EGI58789.1 DS235843 EEB18312.1 KQ977394 KYN02985.1 APGK01017648 APGK01017649 KB740047 KB631866 ENN81834.1 ERL86808.1 GL765434 EFZ16427.1 KQ981856 KYN34588.1 ACPB03006649 GDHC01005061 JAQ13568.1 KQ434879 KZC09982.1 NNAY01000983 OXU25621.1 KQ982557 KYQ55105.1 KQ980624 KYN15033.1 GFTR01008780 JAW07646.1 GFDL01003149 JAV31896.1 ATLV01013242 KE524847 KFB37564.1 JXUM01069058 JXUM01069059 KQ562533 KXJ75685.1 CH477202 EAT48120.1 AAAB01008816 EAA05261.4 AXCM01009566 ADMH02000205 ETN67373.1 JH431841 GGFK01003976 MBW37297.1 LBMM01000214 KMR04458.1 APCN01004490 GAKP01018577 JAC40375.1 GDHF01011246 JAI41068.1 GL453904 EFN75271.1 GBXI01006305 JAD07987.1 KQ976440 KYM86555.1 CVRI01000002 CRK86785.1 CH902619 EDV37560.1 CM000071 EAL25170.2 CH479181 EDW32048.1 CH940648 EDW60602.2 CP012524 ALC41041.1 OUUW01000001 SPP75313.1 CM000158 EDW91032.1 CH954179 EDV56024.1 CH933808 EDW09580.2 CM002911 KMY93778.1 CH480816 EDW47868.1 AF241364 AF241365 AF241366 AE013599 AY051896 CH964282 EDW85820.1 GDRN01103447 JAI58086.1 GEDV01003275 JAP85282.1 CM000362 EDX07094.1 GFPF01006807 MAA17953.1 CH916375 EDV98226.1 CCAG010002095 GEBF01006266 JAN97366.1 KRG00299.1 IACF01002475 LAB68128.1 KN122397 KFO30625.1 JH173457 EHB16887.1 AAQR03179256 AAQR03179257 AAQR03179258 GFFV01000319 JAV39626.1 AC139605 BC168713 AAI68713.1 GBEX01000729 JAI13831.1 AF322193 BC056388 JT406405 AHH37581.1 AQIB01115377 AQIB01115378 X83097 NDHI03003691 PNJ08062.1 PNJ08063.1
NEVH01025648 PNF15214.1 PNF15210.1 KQ761323 OAD57895.1 KZ288309 PBC28675.1 GL434618 EFN74734.1 ADTU01005136 QOIP01000003 RLU24961.1 AAZX01006832 KQ971319 EFA00240.2 GEBQ01030988 JAT08989.1 GECZ01012880 JAS56889.1 GEZM01066160 JAV68121.1 GL888624 EGI58789.1 DS235843 EEB18312.1 KQ977394 KYN02985.1 APGK01017648 APGK01017649 KB740047 KB631866 ENN81834.1 ERL86808.1 GL765434 EFZ16427.1 KQ981856 KYN34588.1 ACPB03006649 GDHC01005061 JAQ13568.1 KQ434879 KZC09982.1 NNAY01000983 OXU25621.1 KQ982557 KYQ55105.1 KQ980624 KYN15033.1 GFTR01008780 JAW07646.1 GFDL01003149 JAV31896.1 ATLV01013242 KE524847 KFB37564.1 JXUM01069058 JXUM01069059 KQ562533 KXJ75685.1 CH477202 EAT48120.1 AAAB01008816 EAA05261.4 AXCM01009566 ADMH02000205 ETN67373.1 JH431841 GGFK01003976 MBW37297.1 LBMM01000214 KMR04458.1 APCN01004490 GAKP01018577 JAC40375.1 GDHF01011246 JAI41068.1 GL453904 EFN75271.1 GBXI01006305 JAD07987.1 KQ976440 KYM86555.1 CVRI01000002 CRK86785.1 CH902619 EDV37560.1 CM000071 EAL25170.2 CH479181 EDW32048.1 CH940648 EDW60602.2 CP012524 ALC41041.1 OUUW01000001 SPP75313.1 CM000158 EDW91032.1 CH954179 EDV56024.1 CH933808 EDW09580.2 CM002911 KMY93778.1 CH480816 EDW47868.1 AF241364 AF241365 AF241366 AE013599 AY051896 CH964282 EDW85820.1 GDRN01103447 JAI58086.1 GEDV01003275 JAP85282.1 CM000362 EDX07094.1 GFPF01006807 MAA17953.1 CH916375 EDV98226.1 CCAG010002095 GEBF01006266 JAN97366.1 KRG00299.1 IACF01002475 LAB68128.1 KN122397 KFO30625.1 JH173457 EHB16887.1 AAQR03179256 AAQR03179257 AAQR03179258 GFFV01000319 JAV39626.1 AC139605 BC168713 AAI68713.1 GBEX01000729 JAI13831.1 AF322193 BC056388 JT406405 AHH37581.1 AQIB01115377 AQIB01115378 X83097 NDHI03003691 PNJ08062.1 PNJ08063.1
Proteomes
UP000005204
UP000218220
UP000027135
UP000235965
UP000242457
UP000005203
+ More
UP000000311 UP000005205 UP000279307 UP000002358 UP000192223 UP000007266 UP000007755 UP000009046 UP000078542 UP000019118 UP000030742 UP000078541 UP000015103 UP000076502 UP000215335 UP000075809 UP000078492 UP000076408 UP000075920 UP000030765 UP000075900 UP000069940 UP000249989 UP000008820 UP000075881 UP000007062 UP000075880 UP000075882 UP000075883 UP000075885 UP000000673 UP000069272 UP000075903 UP000036403 UP000075840 UP000008237 UP000076407 UP000078540 UP000091820 UP000095300 UP000078200 UP000192221 UP000183832 UP000007801 UP000001819 UP000008744 UP000008792 UP000092553 UP000268350 UP000002282 UP000008711 UP000009192 UP000001292 UP000000803 UP000007798 UP000092445 UP000000304 UP000001070 UP000092444 UP000028990 UP000192224 UP000006813 UP000005225 UP000189706 UP000002494 UP000000589 UP000029965 UP000009136
UP000000311 UP000005205 UP000279307 UP000002358 UP000192223 UP000007266 UP000007755 UP000009046 UP000078542 UP000019118 UP000030742 UP000078541 UP000015103 UP000076502 UP000215335 UP000075809 UP000078492 UP000076408 UP000075920 UP000030765 UP000075900 UP000069940 UP000249989 UP000008820 UP000075881 UP000007062 UP000075880 UP000075882 UP000075883 UP000075885 UP000000673 UP000069272 UP000075903 UP000036403 UP000075840 UP000008237 UP000076407 UP000078540 UP000091820 UP000095300 UP000078200 UP000192221 UP000183832 UP000007801 UP000001819 UP000008744 UP000008792 UP000092553 UP000268350 UP000002282 UP000008711 UP000009192 UP000001292 UP000000803 UP000007798 UP000092445 UP000000304 UP000001070 UP000092444 UP000028990 UP000192224 UP000006813 UP000005225 UP000189706 UP000002494 UP000000589 UP000029965 UP000009136
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JWC8
A0A2A4JWZ2
A0A067QMH4
A0A2J7PFU6
A0A2J7PFU9
A0A310SM89
+ More
A0A2A3EAC1 A0A087ZX72 E1ZVR7 A0A158NZY8 A0A3L8DWZ0 K7ITG7 A0A1W4XRH5 D6WFP3 A0A1B6KC09 A0A1B6G369 A0A1Y1L5P4 F4X3Z7 E0VY56 A0A195CQR4 N6TMU2 E9ISL3 A0A195F296 T1HLS2 A0A146M172 A0A154PDP7 A0A232F4C9 A0A151X3V9 A0A151J0U4 A0A224X582 A0A1Q3FWQ1 A0A182Y710 A0A182W1M8 A0A084VHX0 A0A182RLI6 A0A182GK16 Q17N25 A0A182KCV3 Q7QHS8 A0A182INZ6 A0A182LH94 A0A182LV33 A0A1S4H6C5 A0A182P5P9 W5JVI9 A0A182FBZ4 T1J479 A0A182V794 A0A2M4A931 A0A0J7L9D9 A0A182HY45 A0A034VF18 A0A0K8VQE0 E2C9X3 A0A0A1XA51 A0A182WXH0 A0A195BLJ1 A0A1A9W3R0 A0A1I8NUG9 A0A1A9V1N0 A0A1W4U6G1 A0A1J1HKM0 B3ME15 Q291E2 B4GAU6 B4LJU3 A0A0M4E4M5 A0A3B0J601 B4P767 B3NR35 B4KT60 A0A0J9RCL1 B4HRD9 Q9V726 B4NN81 A0A1A9Z1W2 A0A0P4VRN3 A0A131Z3B1 B4QFU9 A0A224YUK0 B4JW88 A0A1B0F9F7 A0A0P6J4F5 A0A0Q9WW52 Q9V726-2 A0A2P2I273 A0A091DK29 A0A1W4XTH9 G5C5S6 H0WG63 A0A1W4Y3B8 A0A1U7QU45 A0A250Y7S1 B5DEL2 A0A0F7ZBG5 Q9EPU4 W5U6H6 A0A0D9R822 Q10569 A0A2J8RHR5
A0A2A3EAC1 A0A087ZX72 E1ZVR7 A0A158NZY8 A0A3L8DWZ0 K7ITG7 A0A1W4XRH5 D6WFP3 A0A1B6KC09 A0A1B6G369 A0A1Y1L5P4 F4X3Z7 E0VY56 A0A195CQR4 N6TMU2 E9ISL3 A0A195F296 T1HLS2 A0A146M172 A0A154PDP7 A0A232F4C9 A0A151X3V9 A0A151J0U4 A0A224X582 A0A1Q3FWQ1 A0A182Y710 A0A182W1M8 A0A084VHX0 A0A182RLI6 A0A182GK16 Q17N25 A0A182KCV3 Q7QHS8 A0A182INZ6 A0A182LH94 A0A182LV33 A0A1S4H6C5 A0A182P5P9 W5JVI9 A0A182FBZ4 T1J479 A0A182V794 A0A2M4A931 A0A0J7L9D9 A0A182HY45 A0A034VF18 A0A0K8VQE0 E2C9X3 A0A0A1XA51 A0A182WXH0 A0A195BLJ1 A0A1A9W3R0 A0A1I8NUG9 A0A1A9V1N0 A0A1W4U6G1 A0A1J1HKM0 B3ME15 Q291E2 B4GAU6 B4LJU3 A0A0M4E4M5 A0A3B0J601 B4P767 B3NR35 B4KT60 A0A0J9RCL1 B4HRD9 Q9V726 B4NN81 A0A1A9Z1W2 A0A0P4VRN3 A0A131Z3B1 B4QFU9 A0A224YUK0 B4JW88 A0A1B0F9F7 A0A0P6J4F5 A0A0Q9WW52 Q9V726-2 A0A2P2I273 A0A091DK29 A0A1W4XTH9 G5C5S6 H0WG63 A0A1W4Y3B8 A0A1U7QU45 A0A250Y7S1 B5DEL2 A0A0F7ZBG5 Q9EPU4 W5U6H6 A0A0D9R822 Q10569 A0A2J8RHR5
PDB
6FUW
E-value=0,
Score=3162
Ontologies
GO
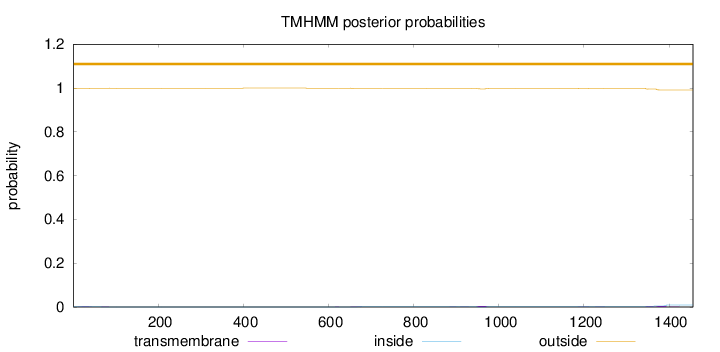
Topology
Subcellular location
Nucleus
Length:
1456
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.347160000000001
Exp number, first 60 AAs:
0.03345
Total prob of N-in:
0.00238
outside
1 - 1456
Population Genetic Test Statistics
Pi
321.567235
Theta
182.022243
Tajima's D
2.998455
CLR
0.146645
CSRT
0.978651067446628
Interpretation
Uncertain
