Gene
KWMTBOMO16394
Pre Gene Modal
BGIBMGA013846
Annotation
PREDICTED:_uncharacterized_protein_LOC106131926_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.196
Sequence
CDS
ATGAATTTTAAAATAATCGTGACGTGGCTCGTTGTTTTTCAAGCGAACGAAATCAGCTGCGACTGTGACAGTTATGTTGAATGTCTTAGTGGTTTCTTCGAGGACTTCTTCTTTGGAGACCCAGAGGAGACGGAAGTGAGAGCGCAAACGTTTAGAACTAACAATACGGAAATCAAAAATATAAATTTCAGTGATCTAGATGCTGTGCTGAAAGATATAACAGAATTTTTGAGTGAAGAAGTTCAAAGTCAGCCGCAGAATGAATCAGATGACGTTTTCAAAACAGTCAAAGCCGAAACGAAAATTTACATTAAGGCATTCGATTTTGGTGCCAACGGTGAAAACGCTGATGGAGCCGAAAGAAAGAATGAAAACTTCGAAGTTGTATCTAAAATACCTGACCGAAATACTAAAGTCCCTATAAATGATGGAACAGCCATAAATGCAAAGTCGAAGAATAGTGATATTATCGAGAAAGGATCAAAACACGTCGGTCAAAATGAGATATTCAATGATGAGCTTTACATGACATATTATGATGATATAGACGACAATTCTACAGATTATCCGGCTATCGACTACGCCGAAATATACAAAGATGTTACTGTTGAAGAACATGTATACATTCCCACACAAGAGCCTTCTATTGATGACTTCGACGGGAATGCTACAAAAAGTGAAAATGATAATCTCATTGAAAATTCTCCGGATAAGTCACGTTCGAAATCAGACGAGGAAAGCGCTGACTTAAATGTAGTTTCCGCATGGATCGTCACTGTGTTTGTGAAAAATACGACCGCGAATGGCCAGTTCGACTATGTGTGCGACGGAGCTCTGATATCGAACCAACATGTGCTGACCGGTGCAAGATGTACAGTCCACGCAAACGCCACGTTCAAGCCTGATGATGTTATAGTTATCGTCGGGAAGAAGTCTTTGCAAGGGTCTGGCGGGAATGAAAAGGCTGTCAGGGTCACCGAAATACACAGACAGGATAATTTCACAATCGTAAATGGTAAAGTTTTAAATGAGTTAGCGGTGCTCAAACTCGATGAGATTGTCTATGTTAATGACGATGTTCAAATCGCGAATATAAGTACAGGTGAACTTGGCGATTCCGACAAATCTTTAACAACTGCATGGGCCTTTTCGGGTGAACTAAAGTCTGTTTACTTCGAGAAAGTAGAAAATGGATCGTGCGCTGACGGCGACTTAGAGAATGTCTTGTGTGCTACTTATGGAAACGAGGTGGCTCTTTGTCCCAGTTACGGAGGGCTCTACGCTGTCAAGGTTGAGAACGCCTGGTTTCTTCGTGGCATCCGATCAGGAGATCCAGCAGAGAGGGGAATTTGCTTCATCAAACGGATCGATTATACAACCTTGGATAATTATTTGGATTGGATCAACCAATTTTTGGAGAAATAA
Protein
MNFKIIVTWLVVFQANEISCDCDSYVECLSGFFEDFFFGDPEETEVRAQTFRTNNTEIKNINFSDLDAVLKDITEFLSEEVQSQPQNESDDVFKTVKAETKIYIKAFDFGANGENADGAERKNENFEVVSKIPDRNTKVPINDGTAINAKSKNSDIIEKGSKHVGQNEIFNDELYMTYYDDIDDNSTDYPAIDYAEIYKDVTVEEHVYIPTQEPSIDDFDGNATKSENDNLIENSPDKSRSKSDEESADLNVVSAWIVTVFVKNTTANGQFDYVCDGALISNQHVLTGARCTVHANATFKPDDVIVIVGKKSLQGSGGNEKAVRVTEIHRQDNFTIVNGKVLNELAVLKLDEIVYVNDDVQIANISTGELGDSDKSLTTAWAFSGELKSVYFEKVENGSCADGDLENVLCATYGNEVALCPSYGGLYAVKVENAWFLRGIRSGDPAERGICFIKRIDYTTLDNYLDWINQFLEK
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
EMBL
Proteomes
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
PDB
1PYT
E-value=8.73962e-10,
Score=153
Ontologies
GO
Topology
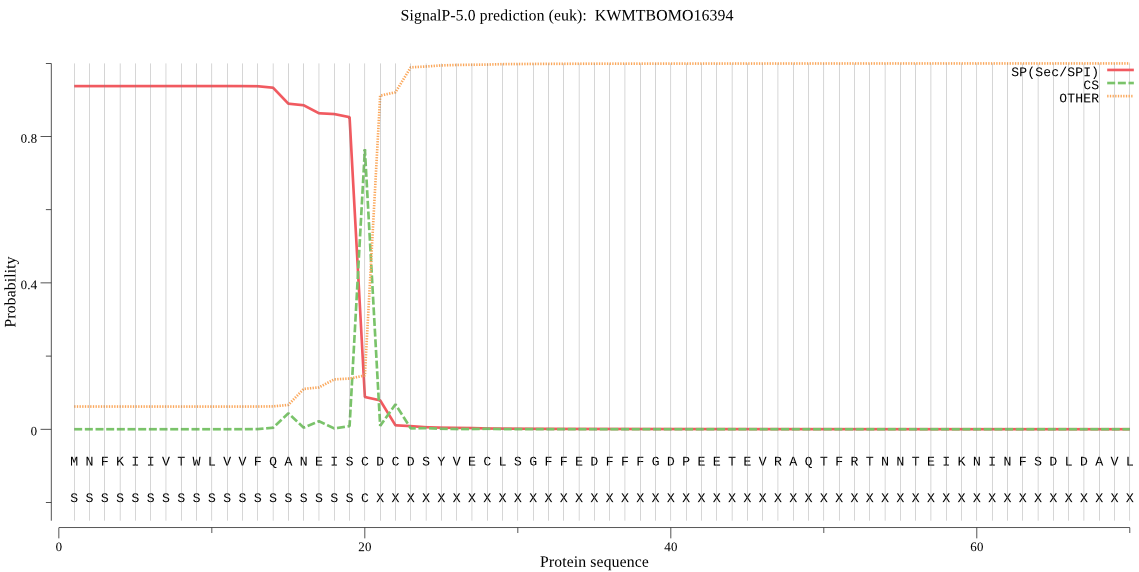
SignalP
Position: 1 - 20,
Likelihood: 0.937516
Length:
474
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00325
Exp number, first 60 AAs:
0.00154
Total prob of N-in:
0.00269
outside
1 - 474
Population Genetic Test Statistics
Pi
224.289965
Theta
183.332206
Tajima's D
1.689872
CLR
11.610114
CSRT
0.826608669566522
Interpretation
Uncertain
