Gene
KWMTBOMO16389
Pre Gene Modal
BGIBMGA013736
Annotation
PREDICTED:_uncharacterized_protein_LOC106142604_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.591
Sequence
CDS
ATGGCAGAAAGAATGCTATTCTTGGAAAAGTTGAAAAAAGCGGCCAGTAAAAATAAAGAAAATGAGGATCAACATGATTTAACGAATAGTGAACAAGAAGTAGAAAGTAATTTACAAATAGAAGCTGAACCAAAAACTATCGAATTGACCAATATTAATGATGAAAGTGAAGAAGATAAGGAAGAAAAAGATTTTTCATCAGATGATAGTATCGCAGATCCTCATTATCAGATTGAAAAGAATGATTTATCGTCAGATTCGGATGAAACAGTAGAACAGAATGAAGAAGAAAATATTTCACGTGAATCAAACCAAAACATTACACGAAGGAGGAAAGGACAGCCAGCACTTTGGAAAACGAATGTAGCAAAGCGTCTACGAAATGCAGGGAAAGAATATGTAGGCAAAAATAACAAAGTTATGCGAGAAAGACGATTAGGACCAGGCTGTACAGAAAAGTGCAAGTTAAAGTGTAACACAAGGTTTGATGATGAGACTCGACGCCATATTTTTGATGCATATTATAAATTACCGGATCTGCAAAGTAAAAGAACTTTTGTAGCATTCAATATGTCCTCTATTGTACCTACGTATCAATATAAAAAAGGAACACGTCGAAATAACAACGCATTTTATTTCAATGATAAAGATGGCGAAAAAATTCGCGTTTGTAAGAGTTTTTTTATGTCTACGCTGGATGTATCTGACAGATTTATTCGAACGGTTACAGAAAAATCAAGAAGTGATGTTTCAGGTATTATACTGGAGGATCAGAGAGGGAAACATGGTAATCACAGCCATGTAAATAATAATGTCAAAGATAATTTGAGAAATTTTATTAAGGCGATACCAAAAATAGAAAGCCATTACTGTCGAGCAGATACGCGAAAAGAATATATTGATGGAAGTAAGACTATTGCAGATATTTATAGAGATTATGTGAAACAATGTGAAGAGAAAAGAGAAACAAGTGCTAATTATATCATGTTTAATAGAATATTTAATGGAGAATTTAATATAGCATTTTACCAACCTAAGAAGGATCAGTGTGAGGACTGTGTTGCCTATAATAACGCAACTGGAGAAGATAAAAACCGACTGAAGCCGAAATATGAAAACCATCTAAAAGAAAAAGAGCTATCAAGAATAGAGAAGGCAAAAGACAAGTCAGACGCAGAAGATAAAGAATCAACTATAGTAGCGGTTTATGACTTACAGGCTGTGATGCCATGTCCTAGGGGAGATGTGTCAAATTTCTACTATGTTTCAAAGGTAAATACCTTAAACTTTACAATCTATGACATGAATAATAAAAATGCGCTCTGCTACGTGTGGCATGAAGGAGAAGCAAACAGAGGAGCTATAGAAATAGGGACCTGTGTTTTAGAATATTTAAAAAATCTTGGCAGGGATTTAACAACTGAAGCGAAACTAGATATTATTTTTTATTCTGACAACTGCTGCGGACAGCAAAAGAATAAGTTTATTATTAGTCTGTACATGTACGCTGTTTCTACTTTTGATTATATCAATAGTATCACGCATAAATTTCTAATAAAAGGTCACACGCAAAACGAAGGTGACAGTGTGCATTCTGTCATTGAAAAAAACATTAAAAAGACTTTGAAATCTGGGCCTATCTATACACCCGATCAGTACATTTCTATAATTCGCACTGCAAAAAAGAAAGGATCTCCTTACAAAGTCATAGAAATGAATCATACAAATTTTTACGATTTAAAATCGCTAGCCTCTGATGTGGGAAGAAATTTTAATAAAAATACTGATAATGAGACTGTGAAGATGACAGATATAAAGATTCTGAAAGTAGATGCTGACCACAAAGGGTTCTTTTTGTATAAAACTTCTTACGGTGAAAATGAATTTAAATTTGTGAAAGTTATTAGCCGAGCATCAAAATCTTTGTACGGAAATTACAGATTGCAATCTGCTTATAAAAAAAGTAACGGAATTTCAGAAAATAAGAAGAGAGGACTACTAGATTTGGTTAACAAAAATATAATTCCAAACTATTATCGACAATTTTACGAAGGGTTATAA
Protein
MAERMLFLEKLKKAASKNKENEDQHDLTNSEQEVESNLQIEAEPKTIELTNINDESEEDKEEKDFSSDDSIADPHYQIEKNDLSSDSDETVEQNEEENISRESNQNITRRRKGQPALWKTNVAKRLRNAGKEYVGKNNKVMRERRLGPGCTEKCKLKCNTRFDDETRRHIFDAYYKLPDLQSKRTFVAFNMSSIVPTYQYKKGTRRNNNAFYFNDKDGEKIRVCKSFFMSTLDVSDRFIRTVTEKSRSDVSGIILEDQRGKHGNHSHVNNNVKDNLRNFIKAIPKIESHYCRADTRKEYIDGSKTIADIYRDYVKQCEEKRETSANYIMFNRIFNGEFNIAFYQPKKDQCEDCVAYNNATGEDKNRLKPKYENHLKEKELSRIEKAKDKSDAEDKESTIVAVYDLQAVMPCPRGDVSNFYYVSKVNTLNFTIYDMNNKNALCYVWHEGEANRGAIEIGTCVLEYLKNLGRDLTTEAKLDIIFYSDNCCGQQKNKFIISLYMYAVSTFDYINSITHKFLIKGHTQNEGDSVHSVIEKNIKKTLKSGPIYTPDQYISIIRTAKKKGSPYKVIEMNHTNFYDLKSLASDVGRNFNKNTDNETVKMTDIKILKVDADHKGFFLYKTSYGENEFKFVKVISRASKSLYGNYRLQSAYKKSNGISENKKRGLLDLVNKNIIPNYYRQFYEGL
Summary
Uniprot
H9JW23
A0A2H1WUL7
J9LMT7
J9KBE8
A0A026WEZ7
T1HH44
+ More
J9KD89 A0A3S2M8H5 A0A0A9VZA8 A0A3S2LBW7 A0A2W1C4W6 A0A2S2NW59 A0A2H1X0U8 A0A0A1XB56 J9LH36 J9LBW7 H9JF09 J9LN28 J9M4G9 J9LYS5 A0A2R7WJY1 X1WUB1 W4ZD48 J9KUZ7 A0A3S2N704 W4XW95 X1XRK2 J9L9L9 A0A3S2P4C8 W4YFK8 V4ACF2 J9LAK0 J9KUZ8 A0A2H1VXX2 J9KRP2 R7USQ6 A0A1Y1MM80 A0A336L7N8 A0A1Y1N893 A0A3S2TF04 J9KF07 J9KUE4 J9M3G1 A0A3N0YTD4 W4XFL6 X1X3M6 J9LY49 J9M2W1 A0A2S2P112 A0A2S2NVW0 X1X655 S4PHD9 W4XFL7 A0A224X9R1 J9M8J2 V4ABT9 A0A1Y1LHW9 V4A9X1 X1XHP1 X1X811 A0A1B0DBG1 A0A3S1ATS4 A0A1B6ERU6 H9JMR7
J9KD89 A0A3S2M8H5 A0A0A9VZA8 A0A3S2LBW7 A0A2W1C4W6 A0A2S2NW59 A0A2H1X0U8 A0A0A1XB56 J9LH36 J9LBW7 H9JF09 J9LN28 J9M4G9 J9LYS5 A0A2R7WJY1 X1WUB1 W4ZD48 J9KUZ7 A0A3S2N704 W4XW95 X1XRK2 J9L9L9 A0A3S2P4C8 W4YFK8 V4ACF2 J9LAK0 J9KUZ8 A0A2H1VXX2 J9KRP2 R7USQ6 A0A1Y1MM80 A0A336L7N8 A0A1Y1N893 A0A3S2TF04 J9KF07 J9KUE4 J9M3G1 A0A3N0YTD4 W4XFL6 X1X3M6 J9LY49 J9M2W1 A0A2S2P112 A0A2S2NVW0 X1X655 S4PHD9 W4XFL7 A0A224X9R1 J9M8J2 V4ABT9 A0A1Y1LHW9 V4A9X1 X1XHP1 X1X811 A0A1B0DBG1 A0A3S1ATS4 A0A1B6ERU6 H9JMR7
EMBL
BABH01025621
ODYU01011180
SOQ56761.1
ABLF02010195
ABLF02010196
ABLF02055224
+ More
ABLF02009832 ABLF02009835 KK107242 EZA54533.1 ACPB03016567 ACPB03016568 ABLF02013766 RSAL01000009 RVE53882.1 GBHO01042097 JAG01507.1 RSAL01000260 RVE43346.1 KZ150145 PZC72904.1 GGMR01008791 MBY21410.1 ODYU01012570 SOQ58975.1 GBXI01005713 JAD08579.1 ABLF02023797 ABLF02019640 ABLF02019643 ABLF02019649 BABH01033861 ABLF02003967 ABLF02022524 ABLF02009837 ABLF02009842 KK854960 PTY19956.1 ABLF02037175 ABLF02037177 AAGJ04019135 AAGJ04019136 AAGJ04019137 ABLF02033230 RSAL01005524 RVE40000.1 AAGJ04159061 ABLF02004032 ABLF02042646 ABLF02004635 RSAL01000569 RVE41377.1 AAGJ04087115 KB200426 ESP01684.1 ABLF02026661 ABLF02026674 ABLF02026675 ABLF02041813 ABLF02060074 ABLF02010095 ODYU01004805 SOQ45064.1 ABLF02013311 AMQN01021160 AMQN01021161 KB298212 ELU09529.1 GEZM01027265 JAV86789.1 UFQS01001897 UFQT01001897 SSX12663.1 SSX32106.1 GEZM01010383 JAV94073.1 RSAL01000210 RVE44232.1 ABLF02020246 ABLF02014818 ABLF02014820 ABLF02006166 ABLF02026282 RJVU01027735 ROL48958.1 AAGJ04162991 ABLF02010023 ABLF02038617 ABLF02012672 GGMR01010491 MBY23110.1 GGMR01008668 MBY21287.1 ABLF02024430 GAIX01003312 JAA89248.1 AAGJ04038612 AAGJ04038613 GFTR01007350 JAW09076.1 ABLF02012895 KB201847 ESO94282.1 GEZM01058056 GEZM01058054 GEZM01058052 GEZM01058051 GEZM01058048 JAV71930.1 KB202199 ESO91835.1 ABLF02018334 ABLF02021172 AJVK01029962 RQTK01001146 RUS71879.1 GECZ01029139 JAS40630.1 BABH01016775
ABLF02009832 ABLF02009835 KK107242 EZA54533.1 ACPB03016567 ACPB03016568 ABLF02013766 RSAL01000009 RVE53882.1 GBHO01042097 JAG01507.1 RSAL01000260 RVE43346.1 KZ150145 PZC72904.1 GGMR01008791 MBY21410.1 ODYU01012570 SOQ58975.1 GBXI01005713 JAD08579.1 ABLF02023797 ABLF02019640 ABLF02019643 ABLF02019649 BABH01033861 ABLF02003967 ABLF02022524 ABLF02009837 ABLF02009842 KK854960 PTY19956.1 ABLF02037175 ABLF02037177 AAGJ04019135 AAGJ04019136 AAGJ04019137 ABLF02033230 RSAL01005524 RVE40000.1 AAGJ04159061 ABLF02004032 ABLF02042646 ABLF02004635 RSAL01000569 RVE41377.1 AAGJ04087115 KB200426 ESP01684.1 ABLF02026661 ABLF02026674 ABLF02026675 ABLF02041813 ABLF02060074 ABLF02010095 ODYU01004805 SOQ45064.1 ABLF02013311 AMQN01021160 AMQN01021161 KB298212 ELU09529.1 GEZM01027265 JAV86789.1 UFQS01001897 UFQT01001897 SSX12663.1 SSX32106.1 GEZM01010383 JAV94073.1 RSAL01000210 RVE44232.1 ABLF02020246 ABLF02014818 ABLF02014820 ABLF02006166 ABLF02026282 RJVU01027735 ROL48958.1 AAGJ04162991 ABLF02010023 ABLF02038617 ABLF02012672 GGMR01010491 MBY23110.1 GGMR01008668 MBY21287.1 ABLF02024430 GAIX01003312 JAA89248.1 AAGJ04038612 AAGJ04038613 GFTR01007350 JAW09076.1 ABLF02012895 KB201847 ESO94282.1 GEZM01058056 GEZM01058054 GEZM01058052 GEZM01058051 GEZM01058048 JAV71930.1 KB202199 ESO91835.1 ABLF02018334 ABLF02021172 AJVK01029962 RQTK01001146 RUS71879.1 GECZ01029139 JAS40630.1 BABH01016775
Proteomes
Pfam
Interpro
IPR018289
MULE_transposase_dom
+ More
IPR007588 Znf_FLYWCH
IPR012337 RNaseH-like_sf
IPR009057 Homeobox-like_sf
IPR007889 HTH_Psq
IPR036388 WH-like_DNA-bd_sf
IPR029526 PGBD
IPR020708 DNA-dir_RNA_polK_14-18kDa_CS
IPR036161 RPB6/omega-like_sf
IPR006110 Pol_omega/K/RPB6
IPR012293 RNAP_RPB6_omega
IPR027806 HARBI1_dom
IPR032071 DUF4806
IPR018247 EF_Hand_1_Ca_BS
IPR007588 Znf_FLYWCH
IPR012337 RNaseH-like_sf
IPR009057 Homeobox-like_sf
IPR007889 HTH_Psq
IPR036388 WH-like_DNA-bd_sf
IPR029526 PGBD
IPR020708 DNA-dir_RNA_polK_14-18kDa_CS
IPR036161 RPB6/omega-like_sf
IPR006110 Pol_omega/K/RPB6
IPR012293 RNAP_RPB6_omega
IPR027806 HARBI1_dom
IPR032071 DUF4806
IPR018247 EF_Hand_1_Ca_BS
Gene 3D
ProteinModelPortal
H9JW23
A0A2H1WUL7
J9LMT7
J9KBE8
A0A026WEZ7
T1HH44
+ More
J9KD89 A0A3S2M8H5 A0A0A9VZA8 A0A3S2LBW7 A0A2W1C4W6 A0A2S2NW59 A0A2H1X0U8 A0A0A1XB56 J9LH36 J9LBW7 H9JF09 J9LN28 J9M4G9 J9LYS5 A0A2R7WJY1 X1WUB1 W4ZD48 J9KUZ7 A0A3S2N704 W4XW95 X1XRK2 J9L9L9 A0A3S2P4C8 W4YFK8 V4ACF2 J9LAK0 J9KUZ8 A0A2H1VXX2 J9KRP2 R7USQ6 A0A1Y1MM80 A0A336L7N8 A0A1Y1N893 A0A3S2TF04 J9KF07 J9KUE4 J9M3G1 A0A3N0YTD4 W4XFL6 X1X3M6 J9LY49 J9M2W1 A0A2S2P112 A0A2S2NVW0 X1X655 S4PHD9 W4XFL7 A0A224X9R1 J9M8J2 V4ABT9 A0A1Y1LHW9 V4A9X1 X1XHP1 X1X811 A0A1B0DBG1 A0A3S1ATS4 A0A1B6ERU6 H9JMR7
J9KD89 A0A3S2M8H5 A0A0A9VZA8 A0A3S2LBW7 A0A2W1C4W6 A0A2S2NW59 A0A2H1X0U8 A0A0A1XB56 J9LH36 J9LBW7 H9JF09 J9LN28 J9M4G9 J9LYS5 A0A2R7WJY1 X1WUB1 W4ZD48 J9KUZ7 A0A3S2N704 W4XW95 X1XRK2 J9L9L9 A0A3S2P4C8 W4YFK8 V4ACF2 J9LAK0 J9KUZ8 A0A2H1VXX2 J9KRP2 R7USQ6 A0A1Y1MM80 A0A336L7N8 A0A1Y1N893 A0A3S2TF04 J9KF07 J9KUE4 J9M3G1 A0A3N0YTD4 W4XFL6 X1X3M6 J9LY49 J9M2W1 A0A2S2P112 A0A2S2NVW0 X1X655 S4PHD9 W4XFL7 A0A224X9R1 J9M8J2 V4ABT9 A0A1Y1LHW9 V4A9X1 X1XHP1 X1X811 A0A1B0DBG1 A0A3S1ATS4 A0A1B6ERU6 H9JMR7
Ontologies
KEGG
GO
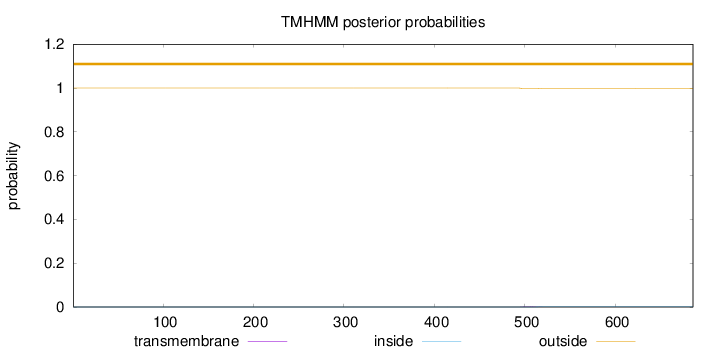
Topology
Length:
686
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02581
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00023
outside
1 - 686
Population Genetic Test Statistics
Pi
1.378872
Theta
4.557019
Tajima's D
-2.341581
CLR
9.727841
CSRT
0.00124993750312484
Interpretation
Uncertain
