Gene
KWMTBOMO16386 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013738
Annotation
PREDICTED:_transmembrane_protease_serine_11A-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.285
Sequence
CDS
ATGTACAGCTTAAGCAAAGTTTTATTCCTCATTGGGATTTTAGTGTGTACGAATGCTAAAGATTATGATAAGCATTGGGCTTTCATTGGGAATCCACTTCTTCATGCAGCTCCGTGCATCGGAAACAGTAAAGTATCTATATCATATGAACCGGGCCTGGCACCGGAGGAAGAAACTAAATACAATTTATTTATAAACGGCCCTTTCCCCGAACACACGACAATCAAAATGAAATTTGATTCGCCCGCCAATGTGACTCTGAGCAACGACATAGGCAAATTAGCAAGAGTGTCTTCGTATGCTGACGGAATGTTCTTCATCAAATACTTTAAAGGCGGACCCTATTTCAGTGCAGATGTGATAGGTTTAAACTTATCCGTAGTACCTTACCCGACCACTATTAACTTAAACTTTGTCGAATACTGCGAGCACCCGGCACTTGGAATCTTAGATGGATATATCAACGGATATAAATCAACAGCTTCATCTACTTATACAGAACACGATGACAACTGTGGAAGAATTAAAATCTCCGAAAAACAATGCGGTGGAAACGAAGAAGAACCTATGCCCTGGCATGCTCTTATCAGAGACGACAACTCTACAATATGCGCTGGAACGTTGATATTACAGAGATATGTTTTGACAGCCGCTCAATGCGTCACAAACTTAGGCGTTGCGAAGAACGCCACAACTCTGTCTGTGGTTCTTGGGAAATTTAATAACACCAGTGATGAATGCTTACAGGAAATGGAGGTGGAAACAGTATTCGTTTACGATGGATACAATTATGAAAATGGAACAAATAACATTGCGTTGTTAGAACTGAAGAACGATGTGGTGTTTAACGAAAAAGTCCAACCTGCGTGTCTGTGGGAGTTCAGCGCATACAAGAAGTTAAATTTGAAAGACATCAAGGGATCTGTCATAAGCACAACTCTCGGCGATGATATTGATGAAGGTCTCGAGGTAGTGAATATGAAAAAGCTTAAGTCTTCTAAAGAAGTAAATGTTTATAGGGAGGAGAAATTTGGATCCAAATTCAACGCCACATCTATCAATCGTAAGTGGAACAATAAAGTTTATGGTAGCTGTCCGTCTACCCACGTGCTTTCAAAGGATTAA
Protein
MYSLSKVLFLIGILVCTNAKDYDKHWAFIGNPLLHAAPCIGNSKVSISYEPGLAPEEETKYNLFINGPFPEHTTIKMKFDSPANVTLSNDIGKLARVSSYADGMFFIKYFKGGPYFSADVIGLNLSVVPYPTTINLNFVEYCEHPALGILDGYINGYKSTASSTYTEHDDNCGRIKISEKQCGGNEEEPMPWHALIRDDNSTICAGTLILQRYVLTAAQCVTNLGVAKNATTLSVVLGKFNNTSDECLQEMEVETVFVYDGYNYENGTNNIALLELKNDVVFNEKVQPACLWEFSAYKKLNLKDIKGSVISTTLGDDIDEGLEVVNMKKLKSSKEVNVYREEKFGSKFNATSINRKWNNKVYGSCPSTHVLSKD
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
EMBL
BABH01025596
BABH01025597
BABH01025598
BABH01025599
BABH01025603
BABH01025604
+ More
BABH01025605 BABH01025606 BABH01025607 BABH01025608 BABH01025609 RSAL01000291 RVE42892.1 KQ460047 KPJ17984.1 KQ459054 KPJ03929.1 AY672791 AAV91013.1 AGBW02012508 OWR44972.1 ODYU01009890 SOQ54607.1 KPJ17983.1 AK402992 BAM19503.1 KPJ03930.1 AGBW02006478 OWR55216.1 AJVK01024995 AJVK01034512 GFDF01005429 JAV08655.1 JTDY01000386 KOB77429.1 AJWK01007837 AJWK01007838
BABH01025605 BABH01025606 BABH01025607 BABH01025608 BABH01025609 RSAL01000291 RVE42892.1 KQ460047 KPJ17984.1 KQ459054 KPJ03929.1 AY672791 AAV91013.1 AGBW02012508 OWR44972.1 ODYU01009890 SOQ54607.1 KPJ17983.1 AK402992 BAM19503.1 KPJ03930.1 AGBW02006478 OWR55216.1 AJVK01024995 AJVK01034512 GFDF01005429 JAV08655.1 JTDY01000386 KOB77429.1 AJWK01007837 AJWK01007838
Proteomes
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
PDB
5JPM
E-value=8.51458e-10,
Score=152
Ontologies
GO
Topology
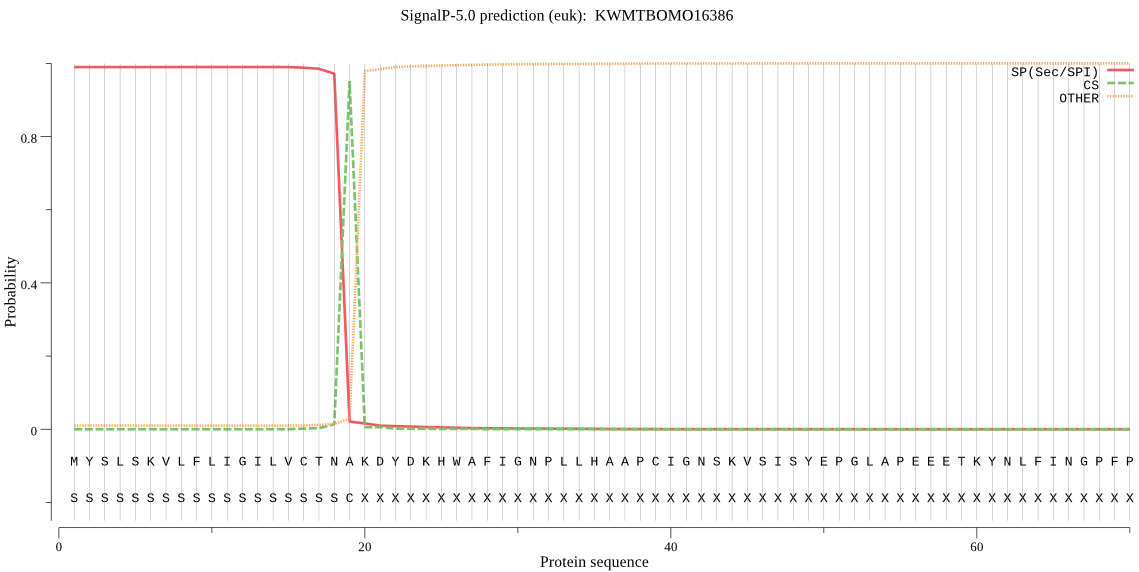
SignalP
Position: 1 - 19,
Likelihood: 0.989727
Length:
374
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04856
Exp number, first 60 AAs:
0.02343
Total prob of N-in:
0.00363
outside
1 - 374

Population Genetic Test Statistics
Pi
413.364018
Theta
173.558005
Tajima's D
3.949325
CLR
0.280743
CSRT
0.998800059997
Interpretation
Uncertain
