Gene
KWMTBOMO16382
Pre Gene Modal
BGIBMGA013740
Annotation
hypothetical_protein_RR48_00843?_partial_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 2.01 Nuclear Reliability : 1.939
Sequence
CDS
ATGTACTGTTTGTTAACAGATGTAGAGTCGATACTTGATTATCACTCAATGCCGGCTCTGTCTGAGCTGGACGACTTCGATCTGTGCCTGAAGAATCCTGAAGCATTGTATTGCATTGTGGACATGGTGCTAATGGAAGATGACACGCCACTGTATCAATTTATTAAGAACTTCTCTTCGTTATCGTACAAAAACTACATGCACTCTAAGCTCCACCGAGGAGTCTGCGCGTCGCAAAACTGTGGACTTAACACAACGTACGCCAATCAAACCGACACCACCGAAAATGAATTGAAGAGATGCTTCAATTCCAGCATACACGAAGGATACGGTCTTCAACAACTATTTGCCAGTTAA
Protein
MYCLLTDVESILDYHSMPALSELDDFDLCLKNPEALYCIVDMVLMEDDTPLYQFIKNFSSLSYKNYMHSKLHRGVCASQNCGLNTTYANQTDTTENELKRCFNSSIHEGYGLQQLFAS
Summary
Uniprot
A0A0N1PG94
A0A2A4JMU9
A0A2H1WCA0
A0A194QKE6
A0A3S2L4H8
M4M6W5
+ More
A0A0L7LPI1 A0A2W1BKB8 A0A3S2P8D1 A0A212EGP9 A0A2H1WAT5 A0A0N0PAP1 A0A194QF64 I4DQA2 A0A2W1BK62 A0A0N1I4J5 A0A212FGA7 A0A0N0P9S1 A0A0L7LAP6 A0A2H1WAR4 A0A194Q1V4 A0A2A4JGE1 A0A194QFD0 A0A2A4JGF7 A0A0N1ID86 A0A0L7KR23 A0A0N1IBJ6 A0A212FM81 A0A194QX14 A0A194QKZ1 A0A0N1PEY2 A0A2H1VKS3 A0A2H1WAT4 A0A194RB57 A0A212ETZ7 A0A0L7L4N3 A0A194QF19 A0A2W1BKH3 A0A182KQ86 A0A2H1WCF8 A0A182N9Y4 A0A194Q6D5 A0A2A4JGW2 H9JU90 A0A2A4JF89 A0A0L7L068 A0A0L7KMG0 B0WNX5 A0A0L7LF62 A0A1Y1K0E5 A0A1Y1K0F7 A0A0L7LK06 A0A1S4FBR5 A0A2A4K7P3 A0A2H1VFZ7
A0A0L7LPI1 A0A2W1BKB8 A0A3S2P8D1 A0A212EGP9 A0A2H1WAT5 A0A0N0PAP1 A0A194QF64 I4DQA2 A0A2W1BK62 A0A0N1I4J5 A0A212FGA7 A0A0N0P9S1 A0A0L7LAP6 A0A2H1WAR4 A0A194Q1V4 A0A2A4JGE1 A0A194QFD0 A0A2A4JGF7 A0A0N1ID86 A0A0L7KR23 A0A0N1IBJ6 A0A212FM81 A0A194QX14 A0A194QKZ1 A0A0N1PEY2 A0A2H1VKS3 A0A2H1WAT4 A0A194RB57 A0A212ETZ7 A0A0L7L4N3 A0A194QF19 A0A2W1BKH3 A0A182KQ86 A0A2H1WCF8 A0A182N9Y4 A0A194Q6D5 A0A2A4JGW2 H9JU90 A0A2A4JF89 A0A0L7L068 A0A0L7KMG0 B0WNX5 A0A0L7LF62 A0A1Y1K0E5 A0A1Y1K0F7 A0A0L7LK06 A0A1S4FBR5 A0A2A4K7P3 A0A2H1VFZ7
EMBL
KQ460527
KPJ14160.1
NWSH01001072
PCG72763.1
ODYU01007576
SOQ50462.1
+ More
KQ459054 KPJ03931.1 RSAL01000159 RVE45485.1 JX846987 AGG55004.1 JTDY01000386 KOB77428.1 KZ150068 PZC74094.1 RSAL01000205 RVE44373.1 AGBW02015045 OWR40663.1 ODYU01007420 SOQ50170.1 KQ461184 KPJ07529.1 KQ459053 KPJ04122.1 AK404010 BAM20092.1 PZC74095.1 KQ459449 KPJ00879.1 AGBW02008687 OWR52781.1 KPJ00884.1 JTDY01002029 KOB72286.1 SOQ50171.1 KQ459582 KPI98974.1 NWSH01001667 PCG70473.1 KPJ04119.1 PCG70472.1 KPJ07535.1 JTDY01006970 KOB65560.1 KPJ07525.1 AGBW02007654 OWR54852.1 KQ461073 KPJ09500.1 KPJ04116.1 KPJ00872.1 ODYU01003108 SOQ41449.1 SOQ50173.1 KQ460436 KPJ14724.1 AGBW02012502 OWR44975.1 JTDY01003022 KOB70291.1 KPJ04014.1 KZ150085 PZC73777.1 SOQ50174.1 KPI98970.1 NWSH01001633 PCG70632.1 BABH01004270 BABH01004271 PCG70631.1 JTDY01004025 KOB68696.1 JTDY01008922 KOB64310.1 DS232017 EDS31994.1 JTDY01001334 KOB74163.1 GEZM01096347 GEZM01096341 JAV54964.1 GEZM01096338 JAV54974.1 JTDY01000850 KOB75674.1 NWSH01000045 PCG80257.1 ODYU01002360 SOQ39759.1
KQ459054 KPJ03931.1 RSAL01000159 RVE45485.1 JX846987 AGG55004.1 JTDY01000386 KOB77428.1 KZ150068 PZC74094.1 RSAL01000205 RVE44373.1 AGBW02015045 OWR40663.1 ODYU01007420 SOQ50170.1 KQ461184 KPJ07529.1 KQ459053 KPJ04122.1 AK404010 BAM20092.1 PZC74095.1 KQ459449 KPJ00879.1 AGBW02008687 OWR52781.1 KPJ00884.1 JTDY01002029 KOB72286.1 SOQ50171.1 KQ459582 KPI98974.1 NWSH01001667 PCG70473.1 KPJ04119.1 PCG70472.1 KPJ07535.1 JTDY01006970 KOB65560.1 KPJ07525.1 AGBW02007654 OWR54852.1 KQ461073 KPJ09500.1 KPJ04116.1 KPJ00872.1 ODYU01003108 SOQ41449.1 SOQ50173.1 KQ460436 KPJ14724.1 AGBW02012502 OWR44975.1 JTDY01003022 KOB70291.1 KPJ04014.1 KZ150085 PZC73777.1 SOQ50174.1 KPI98970.1 NWSH01001633 PCG70632.1 BABH01004270 BABH01004271 PCG70631.1 JTDY01004025 KOB68696.1 JTDY01008922 KOB64310.1 DS232017 EDS31994.1 JTDY01001334 KOB74163.1 GEZM01096347 GEZM01096341 JAV54964.1 GEZM01096338 JAV54974.1 JTDY01000850 KOB75674.1 NWSH01000045 PCG80257.1 ODYU01002360 SOQ39759.1
Proteomes
PRIDE
Pfam
Interpro
IPR002656
Acyl_transf_3
+ More
IPR004119 EcKinase-like
IPR011009 Kinase-like_dom_sf
IPR015897 CHK_kinase-like
IPR020801 PKS_acyl_transferase
IPR016039 Thiolase-like
IPR014030 Ketoacyl_synth_N
IPR020843 PKS_ER
IPR001227 Ac_transferase_dom_sf
IPR029058 AB_hydrolase
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR023102 Fatty_acid_synthase_dom_2
IPR014031 Ketoacyl_synth_C
IPR016035 Acyl_Trfase/lysoPLipase
IPR016036 Malonyl_transacylase_ACP-bd
IPR036291 NAD(P)-bd_dom_sf
IPR014043 Acyl_transferase
IPR001031 Thioesterase
IPR032821 KAsynt_C_assoc
IPR011032 GroES-like_sf
IPR042104 PKS_dehydratase_sf
IPR007577 GlycoTrfase_DXD_sugar-bd_CS
IPR000092 Polyprenyl_synt
IPR039702 FPS1
IPR008949 Isoprenoid_synthase_dom_sf
IPR009003 Peptidase_S1_PA
IPR001254 Trypsin_dom
IPR004119 EcKinase-like
IPR011009 Kinase-like_dom_sf
IPR015897 CHK_kinase-like
IPR020801 PKS_acyl_transferase
IPR016039 Thiolase-like
IPR014030 Ketoacyl_synth_N
IPR020843 PKS_ER
IPR001227 Ac_transferase_dom_sf
IPR029058 AB_hydrolase
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR023102 Fatty_acid_synthase_dom_2
IPR014031 Ketoacyl_synth_C
IPR016035 Acyl_Trfase/lysoPLipase
IPR016036 Malonyl_transacylase_ACP-bd
IPR036291 NAD(P)-bd_dom_sf
IPR014043 Acyl_transferase
IPR001031 Thioesterase
IPR032821 KAsynt_C_assoc
IPR011032 GroES-like_sf
IPR042104 PKS_dehydratase_sf
IPR007577 GlycoTrfase_DXD_sugar-bd_CS
IPR000092 Polyprenyl_synt
IPR039702 FPS1
IPR008949 Isoprenoid_synthase_dom_sf
IPR009003 Peptidase_S1_PA
IPR001254 Trypsin_dom
SUPFAM
ProteinModelPortal
A0A0N1PG94
A0A2A4JMU9
A0A2H1WCA0
A0A194QKE6
A0A3S2L4H8
M4M6W5
+ More
A0A0L7LPI1 A0A2W1BKB8 A0A3S2P8D1 A0A212EGP9 A0A2H1WAT5 A0A0N0PAP1 A0A194QF64 I4DQA2 A0A2W1BK62 A0A0N1I4J5 A0A212FGA7 A0A0N0P9S1 A0A0L7LAP6 A0A2H1WAR4 A0A194Q1V4 A0A2A4JGE1 A0A194QFD0 A0A2A4JGF7 A0A0N1ID86 A0A0L7KR23 A0A0N1IBJ6 A0A212FM81 A0A194QX14 A0A194QKZ1 A0A0N1PEY2 A0A2H1VKS3 A0A2H1WAT4 A0A194RB57 A0A212ETZ7 A0A0L7L4N3 A0A194QF19 A0A2W1BKH3 A0A182KQ86 A0A2H1WCF8 A0A182N9Y4 A0A194Q6D5 A0A2A4JGW2 H9JU90 A0A2A4JF89 A0A0L7L068 A0A0L7KMG0 B0WNX5 A0A0L7LF62 A0A1Y1K0E5 A0A1Y1K0F7 A0A0L7LK06 A0A1S4FBR5 A0A2A4K7P3 A0A2H1VFZ7
A0A0L7LPI1 A0A2W1BKB8 A0A3S2P8D1 A0A212EGP9 A0A2H1WAT5 A0A0N0PAP1 A0A194QF64 I4DQA2 A0A2W1BK62 A0A0N1I4J5 A0A212FGA7 A0A0N0P9S1 A0A0L7LAP6 A0A2H1WAR4 A0A194Q1V4 A0A2A4JGE1 A0A194QFD0 A0A2A4JGF7 A0A0N1ID86 A0A0L7KR23 A0A0N1IBJ6 A0A212FM81 A0A194QX14 A0A194QKZ1 A0A0N1PEY2 A0A2H1VKS3 A0A2H1WAT4 A0A194RB57 A0A212ETZ7 A0A0L7L4N3 A0A194QF19 A0A2W1BKH3 A0A182KQ86 A0A2H1WCF8 A0A182N9Y4 A0A194Q6D5 A0A2A4JGW2 H9JU90 A0A2A4JF89 A0A0L7L068 A0A0L7KMG0 B0WNX5 A0A0L7LF62 A0A1Y1K0E5 A0A1Y1K0F7 A0A0L7LK06 A0A1S4FBR5 A0A2A4K7P3 A0A2H1VFZ7
Ontologies
GO
PANTHER
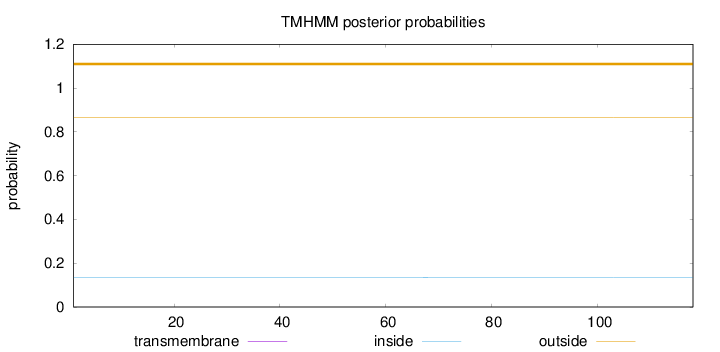
Topology
Length:
118
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00129
Exp number, first 60 AAs:
0.00089
Total prob of N-in:
0.13549
outside
1 - 118
Population Genetic Test Statistics
Pi
48.445434
Theta
42.092038
Tajima's D
1.311949
CLR
3.331559
CSRT
0.747812609369532
Interpretation
Uncertain
