Gene
KWMTBOMO16381
Pre Gene Modal
BGIBMGA013740
Annotation
PREDICTED:_regulator_of_hypoxia-inducible_factor_1-like_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 4.321
Sequence
CDS
ATGGGAGGCTTTTTGATGGCATACAAGATGCTGGAGTATGCTGAAACCCACCGGTTCACCCTTAAAACCGTTCCTATGGCTCTCATTAACCGGTGGTTTAGACTCATTCCAGCAGTTTTAGTCGTGATGGGCCTTGCGATGACTTGGGTACCACATATGGGCTCAGGACCTATGTGGGACGCCGTGGTGAAGAGAGAAAGAGACTTGTGCAGGAAAAACTGGTGGCAATTAGTGATCCTGATGCCCAATCTGTTCCCTTTTGAACATTTATGTCTACCACAAGCTTGGTACTTAGGAACCGATACACAATTATTCTTGATGACATTGGTAATAATGCTGGTCATTTGGCGTTGGCCAAGAAGCGGTGTGCCAGTTCTGTCTGTGGTCATGGCTATTAGTCTTCTTATACCGTTTCTCCAGAGCTATTTCATGAAACTCCTCCCCATCAGAGTCAGCATTTTTCCAGAGTGA
Protein
MGGFLMAYKMLEYAETHRFTLKTVPMALINRWFRLIPAVLVVMGLAMTWVPHMGSGPMWDAVVKRERDLCRKNWWQLVILMPNLFPFEHLCLPQAWYLGTDTQLFLMTLVIMLVIWRWPRSGVPVLSVVMAISLLIPFLQSYFMKLLPIRVSIFPE
Summary
Uniprot
H9JW27
A0A3S2NEL9
A0A2A4J1A0
M4M6W5
A0A0N0PEC2
A0A2H1WCA0
+ More
A0A194QKE6 A0A212EME8 B4NPG1 A0A212ETZ7 A0A2J7QG24 A0A2J7Q6G4 A0A1Y1KVX9 A0A1B0FMU0 A0A212FGA7 B0XH38 A0A182R4K7 A0A1A9VVB4 A0A2W1BKH3 A0A1B0BIV0 A0A1A9Y8R3 A0A1A9Z0F3 A0A1A9X1E4 A0A2A4JEH1 A0A1Y1K6C8 A0A0L7KZR2 A0A1Y1K4D7 A0A3S2LST9 A0A023ESA0 A0A182WLD8 A0A2H1WAT5 Q16PV5 A0A2P8XRP1 A0A2A4J2E4 A0A2H1WAR4 A0A182QX25 A0A2H1WCF8 A0A1Y1K0C4 A0A1Y1K0E4 Q16E87 A0A1B0D242 A0A1Y1K0L9 A0A084WEE0 A0A1Y9GKR7 A0A1Y1K397 A0A1Y1K389 A0A3S2P8D1 A0A1Y9J0V5 A0A1Y1K8A5 A0A182K8B2 A0A182YEG2 A0A139W8X9 A7URR4 A0A1Y9J0F2 A0A182YEG1 A0A1B0CI80 A0A1Y9J0F0 A0A2J7QEN6 A0A2J7QP78 A0A182SXU3 A0A1Y1K4C2 A0A182M972 A0A2H1WFD8 B0WNY8 A0A182U2H9 W8BM55 A0A182W4K6 A0A2J7PZG8 A0A1Y9J0V8 A0A182UXI0 A0A1J1HH25 A0A1W7R5G7 A0A1S4FTS9 A0A2J7QEL1 A0A182L859 A0A182R4J9 A0A182MV59 A0A182PUZ7 A0A1Y9GL72 A0A182JM14 A0A212FDN9 A0A182TJQ2 Q7PTG8 A0A1I8Q0V0 A0A232EIQ2 A7URR5 K7J4D4 A0A2S2QYZ7 A0A1Y1M2Q0 A0A2J7PZF1 A0A182NJC1 A0A2J7PZI3 Q16PV8 Q17BV5 A0A0L7L680 A0A182WLD5 A7URR6 A0A1W7R541
A0A194QKE6 A0A212EME8 B4NPG1 A0A212ETZ7 A0A2J7QG24 A0A2J7Q6G4 A0A1Y1KVX9 A0A1B0FMU0 A0A212FGA7 B0XH38 A0A182R4K7 A0A1A9VVB4 A0A2W1BKH3 A0A1B0BIV0 A0A1A9Y8R3 A0A1A9Z0F3 A0A1A9X1E4 A0A2A4JEH1 A0A1Y1K6C8 A0A0L7KZR2 A0A1Y1K4D7 A0A3S2LST9 A0A023ESA0 A0A182WLD8 A0A2H1WAT5 Q16PV5 A0A2P8XRP1 A0A2A4J2E4 A0A2H1WAR4 A0A182QX25 A0A2H1WCF8 A0A1Y1K0C4 A0A1Y1K0E4 Q16E87 A0A1B0D242 A0A1Y1K0L9 A0A084WEE0 A0A1Y9GKR7 A0A1Y1K397 A0A1Y1K389 A0A3S2P8D1 A0A1Y9J0V5 A0A1Y1K8A5 A0A182K8B2 A0A182YEG2 A0A139W8X9 A7URR4 A0A1Y9J0F2 A0A182YEG1 A0A1B0CI80 A0A1Y9J0F0 A0A2J7QEN6 A0A2J7QP78 A0A182SXU3 A0A1Y1K4C2 A0A182M972 A0A2H1WFD8 B0WNY8 A0A182U2H9 W8BM55 A0A182W4K6 A0A2J7PZG8 A0A1Y9J0V8 A0A182UXI0 A0A1J1HH25 A0A1W7R5G7 A0A1S4FTS9 A0A2J7QEL1 A0A182L859 A0A182R4J9 A0A182MV59 A0A182PUZ7 A0A1Y9GL72 A0A182JM14 A0A212FDN9 A0A182TJQ2 Q7PTG8 A0A1I8Q0V0 A0A232EIQ2 A7URR5 K7J4D4 A0A2S2QYZ7 A0A1Y1M2Q0 A0A2J7PZF1 A0A182NJC1 A0A2J7PZI3 Q16PV8 Q17BV5 A0A0L7L680 A0A182WLD5 A7URR6 A0A1W7R541
Pubmed
EMBL
BABH01025581
RSAL01000159
RVE45486.1
NWSH01003751
PCG65855.1
JX846987
+ More
AGG55004.1 KQ459900 KPJ19352.1 ODYU01007576 SOQ50462.1 KQ459054 KPJ03931.1 AGBW02013856 OWR42649.1 CH964291 EDW86401.2 AGBW02012502 OWR44975.1 NEVH01014836 PNF27544.1 NEVH01017478 PNF24171.1 GEZM01074205 GEZM01074204 JAV64641.1 CCAG010022781 AGBW02008687 OWR52781.1 DS233096 EDS28047.1 KZ150085 PZC73777.1 JXJN01015181 NWSH01001667 PCG70475.1 GEZM01096345 JAV54966.1 JTDY01004025 KOB68697.1 GEZM01096332 JAV54980.1 RSAL01000275 RVE43113.1 GAPW01001480 JAC12118.1 ODYU01007420 SOQ50170.1 CH477771 EAT36393.1 PYGN01001466 PSN34669.1 NWSH01003464 PCG66235.1 SOQ50171.1 AXCN02001261 SOQ50174.1 GEZM01096348 JAV54962.1 GEZM01096333 JAV54979.1 CH478951 EAT32544.1 AJVK01002698 AJVK01002699 AJVK01002700 GEZM01096347 JAV54963.1 ATLV01023205 KE525341 KFB48584.1 APCN01000152 GEZM01096341 JAV54971.1 GEZM01096349 GEZM01096338 GEZM01096336 GEZM01096335 JAV54961.1 RSAL01000205 RVE44373.1 GEZM01096343 JAV54967.1 KQ972879 KXZ75738.1 AAAB01008807 EDO64706.1 AJWK01013083 NEVH01015308 PNF27023.1 NEVH01012096 PNF30392.1 GEZM01096350 JAV54960.1 AXCM01008444 ODYU01008317 SOQ51800.1 DS232017 EDS32007.1 GAMC01012119 JAB94436.1 NEVH01020335 PNF21712.1 CVRI01000004 CRK87301.1 GEHC01001380 JAV46265.1 PNF27022.1 AXCM01001275 AGBW02009017 OWR51851.1 EAA03947.5 NNAY01004216 OXU18191.1 EDO64707.1 GGMS01013773 MBY82976.1 GEZM01042633 JAV79751.1 PNF21711.1 PNF21736.1 EAT36390.1 CH477316 EAT43757.1 JTDY01002748 KOB70794.1 EDO64708.1 GEHC01001381 JAV46264.1
AGG55004.1 KQ459900 KPJ19352.1 ODYU01007576 SOQ50462.1 KQ459054 KPJ03931.1 AGBW02013856 OWR42649.1 CH964291 EDW86401.2 AGBW02012502 OWR44975.1 NEVH01014836 PNF27544.1 NEVH01017478 PNF24171.1 GEZM01074205 GEZM01074204 JAV64641.1 CCAG010022781 AGBW02008687 OWR52781.1 DS233096 EDS28047.1 KZ150085 PZC73777.1 JXJN01015181 NWSH01001667 PCG70475.1 GEZM01096345 JAV54966.1 JTDY01004025 KOB68697.1 GEZM01096332 JAV54980.1 RSAL01000275 RVE43113.1 GAPW01001480 JAC12118.1 ODYU01007420 SOQ50170.1 CH477771 EAT36393.1 PYGN01001466 PSN34669.1 NWSH01003464 PCG66235.1 SOQ50171.1 AXCN02001261 SOQ50174.1 GEZM01096348 JAV54962.1 GEZM01096333 JAV54979.1 CH478951 EAT32544.1 AJVK01002698 AJVK01002699 AJVK01002700 GEZM01096347 JAV54963.1 ATLV01023205 KE525341 KFB48584.1 APCN01000152 GEZM01096341 JAV54971.1 GEZM01096349 GEZM01096338 GEZM01096336 GEZM01096335 JAV54961.1 RSAL01000205 RVE44373.1 GEZM01096343 JAV54967.1 KQ972879 KXZ75738.1 AAAB01008807 EDO64706.1 AJWK01013083 NEVH01015308 PNF27023.1 NEVH01012096 PNF30392.1 GEZM01096350 JAV54960.1 AXCM01008444 ODYU01008317 SOQ51800.1 DS232017 EDS32007.1 GAMC01012119 JAB94436.1 NEVH01020335 PNF21712.1 CVRI01000004 CRK87301.1 GEHC01001380 JAV46265.1 PNF27022.1 AXCM01001275 AGBW02009017 OWR51851.1 EAA03947.5 NNAY01004216 OXU18191.1 EDO64707.1 GGMS01013773 MBY82976.1 GEZM01042633 JAV79751.1 PNF21711.1 PNF21736.1 EAT36390.1 CH477316 EAT43757.1 JTDY01002748 KOB70794.1 EDO64708.1 GEHC01001381 JAV46264.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000007798 UP000235965 UP000092444 UP000002320 UP000075900 UP000078200 UP000092460 UP000092443 UP000092445 UP000091820 UP000037510 UP000075920 UP000008820 UP000245037 UP000075886 UP000092462 UP000030765 UP000075840 UP000076407 UP000075881 UP000076408 UP000007266 UP000007062 UP000092461 UP000075901 UP000075883 UP000075902 UP000075903 UP000183832 UP000075882 UP000075885 UP000075880 UP000095300 UP000215335 UP000002358 UP000075884
UP000007798 UP000235965 UP000092444 UP000002320 UP000075900 UP000078200 UP000092460 UP000092443 UP000092445 UP000091820 UP000037510 UP000075920 UP000008820 UP000245037 UP000075886 UP000092462 UP000030765 UP000075840 UP000076407 UP000075881 UP000076408 UP000007266 UP000007062 UP000092461 UP000075901 UP000075883 UP000075902 UP000075903 UP000183832 UP000075882 UP000075885 UP000075880 UP000095300 UP000215335 UP000002358 UP000075884
PRIDE
Interpro
ProteinModelPortal
H9JW27
A0A3S2NEL9
A0A2A4J1A0
M4M6W5
A0A0N0PEC2
A0A2H1WCA0
+ More
A0A194QKE6 A0A212EME8 B4NPG1 A0A212ETZ7 A0A2J7QG24 A0A2J7Q6G4 A0A1Y1KVX9 A0A1B0FMU0 A0A212FGA7 B0XH38 A0A182R4K7 A0A1A9VVB4 A0A2W1BKH3 A0A1B0BIV0 A0A1A9Y8R3 A0A1A9Z0F3 A0A1A9X1E4 A0A2A4JEH1 A0A1Y1K6C8 A0A0L7KZR2 A0A1Y1K4D7 A0A3S2LST9 A0A023ESA0 A0A182WLD8 A0A2H1WAT5 Q16PV5 A0A2P8XRP1 A0A2A4J2E4 A0A2H1WAR4 A0A182QX25 A0A2H1WCF8 A0A1Y1K0C4 A0A1Y1K0E4 Q16E87 A0A1B0D242 A0A1Y1K0L9 A0A084WEE0 A0A1Y9GKR7 A0A1Y1K397 A0A1Y1K389 A0A3S2P8D1 A0A1Y9J0V5 A0A1Y1K8A5 A0A182K8B2 A0A182YEG2 A0A139W8X9 A7URR4 A0A1Y9J0F2 A0A182YEG1 A0A1B0CI80 A0A1Y9J0F0 A0A2J7QEN6 A0A2J7QP78 A0A182SXU3 A0A1Y1K4C2 A0A182M972 A0A2H1WFD8 B0WNY8 A0A182U2H9 W8BM55 A0A182W4K6 A0A2J7PZG8 A0A1Y9J0V8 A0A182UXI0 A0A1J1HH25 A0A1W7R5G7 A0A1S4FTS9 A0A2J7QEL1 A0A182L859 A0A182R4J9 A0A182MV59 A0A182PUZ7 A0A1Y9GL72 A0A182JM14 A0A212FDN9 A0A182TJQ2 Q7PTG8 A0A1I8Q0V0 A0A232EIQ2 A7URR5 K7J4D4 A0A2S2QYZ7 A0A1Y1M2Q0 A0A2J7PZF1 A0A182NJC1 A0A2J7PZI3 Q16PV8 Q17BV5 A0A0L7L680 A0A182WLD5 A7URR6 A0A1W7R541
A0A194QKE6 A0A212EME8 B4NPG1 A0A212ETZ7 A0A2J7QG24 A0A2J7Q6G4 A0A1Y1KVX9 A0A1B0FMU0 A0A212FGA7 B0XH38 A0A182R4K7 A0A1A9VVB4 A0A2W1BKH3 A0A1B0BIV0 A0A1A9Y8R3 A0A1A9Z0F3 A0A1A9X1E4 A0A2A4JEH1 A0A1Y1K6C8 A0A0L7KZR2 A0A1Y1K4D7 A0A3S2LST9 A0A023ESA0 A0A182WLD8 A0A2H1WAT5 Q16PV5 A0A2P8XRP1 A0A2A4J2E4 A0A2H1WAR4 A0A182QX25 A0A2H1WCF8 A0A1Y1K0C4 A0A1Y1K0E4 Q16E87 A0A1B0D242 A0A1Y1K0L9 A0A084WEE0 A0A1Y9GKR7 A0A1Y1K397 A0A1Y1K389 A0A3S2P8D1 A0A1Y9J0V5 A0A1Y1K8A5 A0A182K8B2 A0A182YEG2 A0A139W8X9 A7URR4 A0A1Y9J0F2 A0A182YEG1 A0A1B0CI80 A0A1Y9J0F0 A0A2J7QEN6 A0A2J7QP78 A0A182SXU3 A0A1Y1K4C2 A0A182M972 A0A2H1WFD8 B0WNY8 A0A182U2H9 W8BM55 A0A182W4K6 A0A2J7PZG8 A0A1Y9J0V8 A0A182UXI0 A0A1J1HH25 A0A1W7R5G7 A0A1S4FTS9 A0A2J7QEL1 A0A182L859 A0A182R4J9 A0A182MV59 A0A182PUZ7 A0A1Y9GL72 A0A182JM14 A0A212FDN9 A0A182TJQ2 Q7PTG8 A0A1I8Q0V0 A0A232EIQ2 A7URR5 K7J4D4 A0A2S2QYZ7 A0A1Y1M2Q0 A0A2J7PZF1 A0A182NJC1 A0A2J7PZI3 Q16PV8 Q17BV5 A0A0L7L680 A0A182WLD5 A7URR6 A0A1W7R541
Ontologies
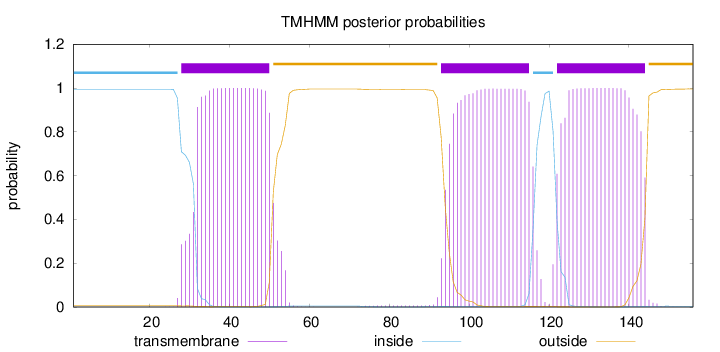
Topology
Length:
156
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
65.27072
Exp number, first 60 AAs:
21.31161
Total prob of N-in:
0.99476
POSSIBLE N-term signal
sequence
inside
1 - 27
TMhelix
28 - 50
outside
51 - 92
TMhelix
93 - 115
inside
116 - 121
TMhelix
122 - 144
outside
145 - 156
Population Genetic Test Statistics
Pi
258.88994
Theta
175.337576
Tajima's D
1.779383
CLR
0.060411
CSRT
0.83900804959752
Interpretation
Uncertain
