Gene
KWMTBOMO16380
Pre Gene Modal
BGIBMGA013740
Annotation
Nose_resistant_to_fluoxetine_protein_6_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.765
Sequence
CDS
ATGGTTACATCCATAAGAGACATATTCGGTTACAATGACACATTCTATCACGCCTACGTGTCAGCGCAAGGAAATTGGGCCGGTTACCATTTAGGAGTTCTCACCGCTTATTTCTATAGAAAAGCCCAGAACCAAAATTGGGATTTGGGCAACTCTTTTCTAATGAGACTTCTCTTCATAGCTTCAGTACCAATAGCTCTAGGTACTGTCCTAATGGGCTGGGATCTACACCACCGCGAAGCTTCGGTCTTTGAATCAGCGATCTTCAACGCTCTCAATCAGAACTTTTTCGCTTTGGCCATGTGTGTCTTCTTTATTGGATACTTCTTCAGATGTAATAAGATTTACGTAAGCGCGGTAGAGTGGGGTCCCTTGCAACCCTTGGGAAGGATGTCTTACTGCGCCATGTTGCTTCACGCCACGATTTTGAGGACCTACGGTGGTCAGATGAGACGGTCTTTCTATGCTACTGATTACACAGCTATTATGCTTTACGCCGGAATCGTAAGCACCACGTATTTATTCGCTCTGCCACTGTACTTATTCGTTGAAGCACCCGCCTGCCAACTTCAGAAGCTCCTTCTGGGCCCAAAAAGACGGATACAGCAAAAAGAAGACCTCAATGCTAACCAAGTGAAGCCTGGAATATCTAATGTCTCCGTATCTACAGTTTCTACGCATATATGA
Protein
MVTSIRDIFGYNDTFYHAYVSAQGNWAGYHLGVLTAYFYRKAQNQNWDLGNSFLMRLLFIASVPIALGTVLMGWDLHHREASVFESAIFNALNQNFFALAMCVFFIGYFFRCNKIYVSAVEWGPLQPLGRMSYCAMLLHATILRTYGGQMRRSFYATDYTAIMLYAGIVSTTYLFALPLYLFVEAPACQLQKLLLGPKRRIQQKEDLNANQVKPGISNVSVSTVSTHI
Summary
Catalytic Activity
S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine + [acceptor protein]-L-lysine = [E2 ubiquitin-conjugating enzyme]-L-cysteine + N(6)-ubiquitinyl-[acceptor protein]-L-lysine.
Uniprot
H9JW27
A0A2A4J1A0
A0A2H1WCA0
M4M6W5
A0A3S2NEL9
A0A194QKE6
+ More
A0A0N0PEC2 A0A084WED7 A0A0N1I9M0 A0A1Y1KVX9 A0A084WED6 A0A194QX14 A0A194QF68 A0A194QFD0 A0A194QVJ9 A0A2H1WAT5 A0A2A4JEH1 W5JEY9 H9JSX5 A0A212FGA7 A0A1Y1LVK7 A0A212EPG8 A0A1Y1K3A0 A0A1Y1K4D7 A0A1Y1K0E4 A0A1Y1K389 A0A1Y1K8A5 A0A1Y1K5L3 A0A1Y1K4C2 A0A1Y1K397 A0A1Y1K0C4 A0A1Y1K399 A0A2A4JGF7 A0A1Y1K0D5 A7URR4 A0A1Y9GL72 A0A1Y9GKS0 A0A2W1BMJ8 A0A2H1WCF8 A0A1B6DTW6 A0A182QFB6 A0A182J629 A0A2H1WAR4 A0A084WEE0 A0A212EME8 A0A3S2P8D1 A0A182TJQ2 A0A1Y9J0F2 A0A1Y1K0D8 A0A1Y1M2Q0 A7URR7 A0A194Q6D5 A0A182KQ86 A0A0N0PCP5 Q7PTG8 A0A1Y9GKR7 A0A087T4D3 A0A182UXI0 W5JRK7 W5JVJ3 A0A0A9X1V1 A0A1S4FTS9 A0A0A9WNL8 A0A194QEZ9 B0WNY8 A0A182QA40 A0A182I2D8 A0A182YEG4 A0A3S2LZ61 I4DNS7 A0A0L7L068 B4M0Z1 Q16PV8 A0A182FHE0 A0A0A9X7B5 A0A2S2NXW1 A0A0A1X2L2 J9KT87 A0A182JNH6 A0A182NJB8 A0NBK6 A0A182FHD9 A0A182WLD8 A0A0L7KZR2 A0A182KQ81 A0A212EGP9 A0A182R1B6 A0A182IST2 A0A1Y9J0V8 A0A087T4D7 A7URR6 A0A182MV59 A0A210QXD8 B4JFD1 A0A084WED5
A0A0N0PEC2 A0A084WED7 A0A0N1I9M0 A0A1Y1KVX9 A0A084WED6 A0A194QX14 A0A194QF68 A0A194QFD0 A0A194QVJ9 A0A2H1WAT5 A0A2A4JEH1 W5JEY9 H9JSX5 A0A212FGA7 A0A1Y1LVK7 A0A212EPG8 A0A1Y1K3A0 A0A1Y1K4D7 A0A1Y1K0E4 A0A1Y1K389 A0A1Y1K8A5 A0A1Y1K5L3 A0A1Y1K4C2 A0A1Y1K397 A0A1Y1K0C4 A0A1Y1K399 A0A2A4JGF7 A0A1Y1K0D5 A7URR4 A0A1Y9GL72 A0A1Y9GKS0 A0A2W1BMJ8 A0A2H1WCF8 A0A1B6DTW6 A0A182QFB6 A0A182J629 A0A2H1WAR4 A0A084WEE0 A0A212EME8 A0A3S2P8D1 A0A182TJQ2 A0A1Y9J0F2 A0A1Y1K0D8 A0A1Y1M2Q0 A7URR7 A0A194Q6D5 A0A182KQ86 A0A0N0PCP5 Q7PTG8 A0A1Y9GKR7 A0A087T4D3 A0A182UXI0 W5JRK7 W5JVJ3 A0A0A9X1V1 A0A1S4FTS9 A0A0A9WNL8 A0A194QEZ9 B0WNY8 A0A182QA40 A0A182I2D8 A0A182YEG4 A0A3S2LZ61 I4DNS7 A0A0L7L068 B4M0Z1 Q16PV8 A0A182FHE0 A0A0A9X7B5 A0A2S2NXW1 A0A0A1X2L2 J9KT87 A0A182JNH6 A0A182NJB8 A0NBK6 A0A182FHD9 A0A182WLD8 A0A0L7KZR2 A0A182KQ81 A0A212EGP9 A0A182R1B6 A0A182IST2 A0A1Y9J0V8 A0A087T4D7 A7URR6 A0A182MV59 A0A210QXD8 B4JFD1 A0A084WED5
Pubmed
EMBL
BABH01025581
NWSH01003751
PCG65855.1
ODYU01007576
SOQ50462.1
JX846987
+ More
AGG55004.1 RSAL01000159 RVE45486.1 KQ459054 KPJ03931.1 KQ459900 KPJ19352.1 ATLV01023204 KE525341 KFB48581.1 KQ459449 KPJ00868.1 GEZM01074205 GEZM01074204 JAV64641.1 KFB48580.1 KQ461073 KPJ09500.1 KQ459053 KPJ04127.1 KPJ04119.1 KPJ09497.1 ODYU01007420 SOQ50170.1 NWSH01001667 PCG70475.1 ADMH02001404 ETN62656.1 BABH01038932 AGBW02008687 OWR52781.1 GEZM01045934 JAV77613.1 AGBW02013474 OWR43366.1 GEZM01096331 JAV54981.1 GEZM01096332 JAV54980.1 GEZM01096333 JAV54979.1 GEZM01096349 GEZM01096338 GEZM01096336 GEZM01096335 JAV54961.1 GEZM01096343 JAV54967.1 GEZM01096346 JAV54965.1 GEZM01096350 JAV54960.1 GEZM01096341 JAV54971.1 GEZM01096348 JAV54962.1 GEZM01096342 JAV54968.1 PCG70472.1 GEZM01096340 JAV54972.1 AAAB01008807 EDO64706.1 APCN01000152 APCN01000148 APCN01000149 KZ150068 PZC74096.1 SOQ50174.1 GEDC01008190 JAS29108.1 AXCN02000449 SOQ50171.1 ATLV01023205 KFB48584.1 AGBW02013856 OWR42649.1 RSAL01000205 RVE44373.1 GEZM01096330 JAV54982.1 GEZM01042633 JAV79751.1 EDO64709.1 KQ459582 KPI98970.1 KQ460635 KPJ13420.1 EAA03947.5 KK113363 KFM59972.1 ADMH02000306 ETN67037.1 ETN67035.1 GBHO01028912 JAG14692.1 GBHO01037154 GBRD01003400 JAG06450.1 JAG62421.1 KPJ04118.1 DS232017 EDS32007.1 APCN01000146 RSAL01000102 RVE47464.1 AK403118 BAM19567.1 JTDY01004025 KOB68696.1 CH940650 EDW67402.2 CH477771 EAT36390.1 GBHO01028911 JAG14693.1 GGMR01009283 MBY21902.1 GBXI01009399 JAD04893.1 ABLF02022069 EAU77672.2 KOB68697.1 AGBW02015045 OWR40663.1 AXCN02000448 KFM59976.1 EDO64708.1 AXCM01001275 NEDP02001363 OWF53373.1 CH916369 EDV93412.1 ATLV01023202 ATLV01023203 KFB48579.1
AGG55004.1 RSAL01000159 RVE45486.1 KQ459054 KPJ03931.1 KQ459900 KPJ19352.1 ATLV01023204 KE525341 KFB48581.1 KQ459449 KPJ00868.1 GEZM01074205 GEZM01074204 JAV64641.1 KFB48580.1 KQ461073 KPJ09500.1 KQ459053 KPJ04127.1 KPJ04119.1 KPJ09497.1 ODYU01007420 SOQ50170.1 NWSH01001667 PCG70475.1 ADMH02001404 ETN62656.1 BABH01038932 AGBW02008687 OWR52781.1 GEZM01045934 JAV77613.1 AGBW02013474 OWR43366.1 GEZM01096331 JAV54981.1 GEZM01096332 JAV54980.1 GEZM01096333 JAV54979.1 GEZM01096349 GEZM01096338 GEZM01096336 GEZM01096335 JAV54961.1 GEZM01096343 JAV54967.1 GEZM01096346 JAV54965.1 GEZM01096350 JAV54960.1 GEZM01096341 JAV54971.1 GEZM01096348 JAV54962.1 GEZM01096342 JAV54968.1 PCG70472.1 GEZM01096340 JAV54972.1 AAAB01008807 EDO64706.1 APCN01000152 APCN01000148 APCN01000149 KZ150068 PZC74096.1 SOQ50174.1 GEDC01008190 JAS29108.1 AXCN02000449 SOQ50171.1 ATLV01023205 KFB48584.1 AGBW02013856 OWR42649.1 RSAL01000205 RVE44373.1 GEZM01096330 JAV54982.1 GEZM01042633 JAV79751.1 EDO64709.1 KQ459582 KPI98970.1 KQ460635 KPJ13420.1 EAA03947.5 KK113363 KFM59972.1 ADMH02000306 ETN67037.1 ETN67035.1 GBHO01028912 JAG14692.1 GBHO01037154 GBRD01003400 JAG06450.1 JAG62421.1 KPJ04118.1 DS232017 EDS32007.1 APCN01000146 RSAL01000102 RVE47464.1 AK403118 BAM19567.1 JTDY01004025 KOB68696.1 CH940650 EDW67402.2 CH477771 EAT36390.1 GBHO01028911 JAG14693.1 GGMR01009283 MBY21902.1 GBXI01009399 JAD04893.1 ABLF02022069 EAU77672.2 KOB68697.1 AGBW02015045 OWR40663.1 AXCN02000448 KFM59976.1 EDO64708.1 AXCM01001275 NEDP02001363 OWF53373.1 CH916369 EDV93412.1 ATLV01023202 ATLV01023203 KFB48579.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000030765
+ More
UP000000673 UP000007151 UP000007062 UP000075840 UP000075886 UP000075880 UP000075902 UP000076407 UP000075882 UP000054359 UP000075903 UP000002320 UP000076408 UP000037510 UP000008792 UP000008820 UP000069272 UP000007819 UP000075881 UP000075884 UP000075920 UP000075883 UP000242188 UP000001070
UP000000673 UP000007151 UP000007062 UP000075840 UP000075886 UP000075880 UP000075902 UP000076407 UP000075882 UP000054359 UP000075903 UP000002320 UP000076408 UP000037510 UP000008792 UP000008820 UP000069272 UP000007819 UP000075881 UP000075884 UP000075920 UP000075883 UP000242188 UP000001070
Interpro
Gene 3D
ProteinModelPortal
H9JW27
A0A2A4J1A0
A0A2H1WCA0
M4M6W5
A0A3S2NEL9
A0A194QKE6
+ More
A0A0N0PEC2 A0A084WED7 A0A0N1I9M0 A0A1Y1KVX9 A0A084WED6 A0A194QX14 A0A194QF68 A0A194QFD0 A0A194QVJ9 A0A2H1WAT5 A0A2A4JEH1 W5JEY9 H9JSX5 A0A212FGA7 A0A1Y1LVK7 A0A212EPG8 A0A1Y1K3A0 A0A1Y1K4D7 A0A1Y1K0E4 A0A1Y1K389 A0A1Y1K8A5 A0A1Y1K5L3 A0A1Y1K4C2 A0A1Y1K397 A0A1Y1K0C4 A0A1Y1K399 A0A2A4JGF7 A0A1Y1K0D5 A7URR4 A0A1Y9GL72 A0A1Y9GKS0 A0A2W1BMJ8 A0A2H1WCF8 A0A1B6DTW6 A0A182QFB6 A0A182J629 A0A2H1WAR4 A0A084WEE0 A0A212EME8 A0A3S2P8D1 A0A182TJQ2 A0A1Y9J0F2 A0A1Y1K0D8 A0A1Y1M2Q0 A7URR7 A0A194Q6D5 A0A182KQ86 A0A0N0PCP5 Q7PTG8 A0A1Y9GKR7 A0A087T4D3 A0A182UXI0 W5JRK7 W5JVJ3 A0A0A9X1V1 A0A1S4FTS9 A0A0A9WNL8 A0A194QEZ9 B0WNY8 A0A182QA40 A0A182I2D8 A0A182YEG4 A0A3S2LZ61 I4DNS7 A0A0L7L068 B4M0Z1 Q16PV8 A0A182FHE0 A0A0A9X7B5 A0A2S2NXW1 A0A0A1X2L2 J9KT87 A0A182JNH6 A0A182NJB8 A0NBK6 A0A182FHD9 A0A182WLD8 A0A0L7KZR2 A0A182KQ81 A0A212EGP9 A0A182R1B6 A0A182IST2 A0A1Y9J0V8 A0A087T4D7 A7URR6 A0A182MV59 A0A210QXD8 B4JFD1 A0A084WED5
A0A0N0PEC2 A0A084WED7 A0A0N1I9M0 A0A1Y1KVX9 A0A084WED6 A0A194QX14 A0A194QF68 A0A194QFD0 A0A194QVJ9 A0A2H1WAT5 A0A2A4JEH1 W5JEY9 H9JSX5 A0A212FGA7 A0A1Y1LVK7 A0A212EPG8 A0A1Y1K3A0 A0A1Y1K4D7 A0A1Y1K0E4 A0A1Y1K389 A0A1Y1K8A5 A0A1Y1K5L3 A0A1Y1K4C2 A0A1Y1K397 A0A1Y1K0C4 A0A1Y1K399 A0A2A4JGF7 A0A1Y1K0D5 A7URR4 A0A1Y9GL72 A0A1Y9GKS0 A0A2W1BMJ8 A0A2H1WCF8 A0A1B6DTW6 A0A182QFB6 A0A182J629 A0A2H1WAR4 A0A084WEE0 A0A212EME8 A0A3S2P8D1 A0A182TJQ2 A0A1Y9J0F2 A0A1Y1K0D8 A0A1Y1M2Q0 A7URR7 A0A194Q6D5 A0A182KQ86 A0A0N0PCP5 Q7PTG8 A0A1Y9GKR7 A0A087T4D3 A0A182UXI0 W5JRK7 W5JVJ3 A0A0A9X1V1 A0A1S4FTS9 A0A0A9WNL8 A0A194QEZ9 B0WNY8 A0A182QA40 A0A182I2D8 A0A182YEG4 A0A3S2LZ61 I4DNS7 A0A0L7L068 B4M0Z1 Q16PV8 A0A182FHE0 A0A0A9X7B5 A0A2S2NXW1 A0A0A1X2L2 J9KT87 A0A182JNH6 A0A182NJB8 A0NBK6 A0A182FHD9 A0A182WLD8 A0A0L7KZR2 A0A182KQ81 A0A212EGP9 A0A182R1B6 A0A182IST2 A0A1Y9J0V8 A0A087T4D7 A7URR6 A0A182MV59 A0A210QXD8 B4JFD1 A0A084WED5
Ontologies
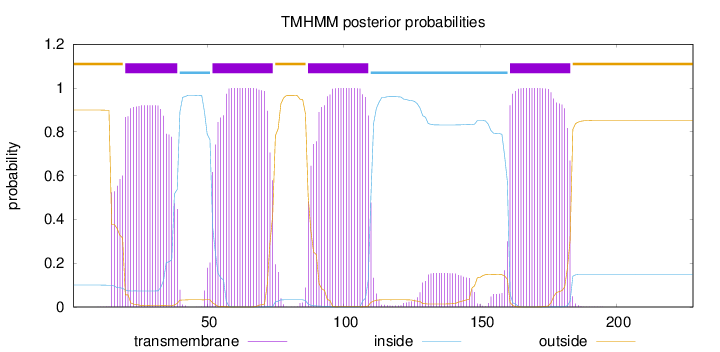
Topology
Length:
228
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
89.5231300000001
Exp number, first 60 AAs:
28.15368
Total prob of N-in:
0.10008
POSSIBLE N-term signal
sequence
outside
1 - 19
TMhelix
20 - 39
inside
40 - 51
TMhelix
52 - 74
outside
75 - 86
TMhelix
87 - 109
inside
110 - 160
TMhelix
161 - 183
outside
184 - 228
Population Genetic Test Statistics
Pi
221.788911
Theta
138.032418
Tajima's D
2.060794
CLR
0.031435
CSRT
0.894705264736763
Interpretation
Uncertain
