Gene
KWMTBOMO16376
Pre Gene Modal
BGIBMGA013742
Annotation
PREDICTED:_PRKCA-binding_protein_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.192
Sequence
CDS
ATGATGCAAGAATATGAAGACGACTTTTTATTTGAAGAGGATAAAATGGGTATGATCGTGACGTCTGGTAGCGTCGTGCTGAGTAAAGATGAAAAGAACTTGATAGGTATCAGCATCGGTGGTGGAGCTCCATTGTGTCCATGCCTTTACATCGTACAGATATTCGACAACACTCCAGCATCCAAGGAAGGCACCCTGCAGAGCGGTGACGAGCTGGTCGGTGTCAATGGGCAGTCCGTAAAAGGAAAAACCAAAGTTGAGGTCGCGAAAATGATACAGTCGGCCAAGGATGAAGTAACAATAAACTATAACAAACTCCACGCCGATCCCAAACAAGGGAAAAGTCTAGATATAATCATGAAGAAGATGAAACACAGATTAGTAGAGAACATGTCTTCGGGGGCCGCTGATGCCCTTGGATTATCGAGGGCTATACTGTGCAATGACACGCTGGTAGCGAAACTGGACGAGCTGAGGGAAACCGAGAATACATACAAGAGACTCGTGGAGCATTCTAAGAGAATGTTAAAGTCGTACTTTGACTTACTCCAAACTTACAAAGCTGTCGGGGATATATTCTCGGCGATCGGGGTGAGAGAGCCTCAGGCTAGAGCTTCCGAGGCTTTCTCGAAGTTCGGTCAATATCATAGACTGCTCGAACGAGACGGCATTAAAATGTTGAAAACTCTAAAGCCGATCCTGTCTGACATGGGCACTTACTTGAACAAAGCTATACCGGACACCAAATTGACGATAAGGAAATACGCGGACACGAAATTCGAGTATCTGTCCTACTGTCTGAAGGTCAAAGAGATGGATGACGAGGAATACGGGTATAACGCTCTGCAGGAGCCTCTGTACCGAGTGGAAACCGGGAATTATGAATATAGGTTGATATTGCGTTGTCGCCAAGACGCCAGAACAAGGTTCGCCAGACTCAGGTCGGACGTTCTGGTAAAGCTGGAGCTTCTAGAGAATAAGAAAGCGCAGGACGTCGCCCATCAACTCAAGAAGTTCATTCAGGGACTGGCCGTTTACCACAACGAAACCTTGGAGCATTTAATGGAGAATTCTACATTGTTCCCAGTCGAAGTTGACCTCTCGCAGAACTGTTTTCAGTATAAATCGGCAGGTCAAATTAGTGAGGACGTCCAAGATGAAGATATAGAATACGCAAAAGAAATACAAGAGTACCACGATTTGTCTGTTAACGACACGAGAGAGGCGAAGTTGATGAAGAAACGTCAGACGGACGACAATGAGCACAATGACGCTACCGAACAGCTTCTCCCGGGTATCGACACCGTTGACATTGACAGGTCCGAGAATAATGACGTTTCGGACAACTTGACACTCCTGACAGAGCTAGGGCTGGCGGACGCGGGAGCCAATGACGAATTCGGGAGCTTCCAAAACGGACTGATGGACGATTTCATGCCGAAGAGCAGCAGCGACGTCGATGACGTTTTCGATAAGCTGTTGAACGATTTGAGTATCGGAAAATAA
Protein
MMQEYEDDFLFEEDKMGMIVTSGSVVLSKDEKNLIGISIGGGAPLCPCLYIVQIFDNTPASKEGTLQSGDELVGVNGQSVKGKTKVEVAKMIQSAKDEVTINYNKLHADPKQGKSLDIIMKKMKHRLVENMSSGAADALGLSRAILCNDTLVAKLDELRETENTYKRLVEHSKRMLKSYFDLLQTYKAVGDIFSAIGVREPQARASEAFSKFGQYHRLLERDGIKMLKTLKPILSDMGTYLNKAIPDTKLTIRKYADTKFEYLSYCLKVKEMDDEEYGYNALQEPLYRVETGNYEYRLILRCRQDARTRFARLRSDVLVKLELLENKKAQDVAHQLKKFIQGLAVYHNETLEHLMENSTLFPVEVDLSQNCFQYKSAGQISEDVQDEDIEYAKEIQEYHDLSVNDTREAKLMKKRQTDDNEHNDATEQLLPGIDTVDIDRSENNDVSDNLTLLTELGLADAGANDEFGSFQNGLMDDFMPKSSSDVDDVFDKLLNDLSIGK
Summary
Uniprot
H9JW29
A0A2H1W120
A0A1E1WTG6
A0A2A4JWZ9
A0A2A4JYG0
A0A1E1WU61
+ More
E0AE01 A0A212EXH0 A0A194QEU0 Q179Z8 A0A1S4GX24 A0A2J7Q1Z0 A0A084VUJ5 B0WEC7 A0A2J7Q1Y3 A0A182JDB4 A0A182IEK3 Q7Q3E3 A0A182X1M3 A0A182L2E6 A0A182TQV1 A0A182UTW7 A0A2J7Q1X9 A0A182F2Z9 A0A182QR49 A0A2J7Q1X8 A0A182JZU0 A0A182R3C0 A0A0A9WUN8 A0A182MFB7 A0A1Q3FYV3 A0A182GRF1 A0A182MZF5 A0A182WA31 A0A182SAY4 A0A0A9X221 A0A1B6CB83 A0A2P8YHA5 A0A1W4XUI1 A0A146M315 D6W6B9 A0A1B6E8U1 A0A131YVU5 A0A1B6CT37 N6UD82 E0VGF8 A0A023GCL6 A0A1W4UTJ4 A0A0J9R1G1 A0A182YGR6 Q8SY07 A0A0P9A5N5 B3N3D2 A0A1B6D349 A0A0R1DMD6 A0A1Y1N2C8 A0A1L8DJE2 A0A1L8DM79 A0A2R5LDK8 A0A1W4XTL2 V9ICV7 A0A293MTF9 A0A1W4XJH4 A0A0R3NTZ0 A0A131XNL6 A0A1B6CG67 A0A1V9XUE6 A0A0Q9X3U2 K7IYN3 A0A1B6D7R3 A0A0C9RVG5 A0A0A1WVU7 A0A3S3NWF1 W8C1N0 A0A147BTM3 A0A034VQN1 B4KIN4 A0A0K8V8K8 A0A0L7QNV3 A0A1W4V5W7 I6YMZ0 B3MNJ3 B4Q486 A0A0J9TM68 X2JDZ1 A0A0P4XQH1 B4MDL9 A0A0P4XJ46 A0A0M5J3W9 A0A0J9R150 A0A3S0ZXP0 A0A0Q5WJK4 V4AYC5 A0A2C9JFN1 A0A0M8ZNU5 A0A026WK07 T1FPB1 A0A0L8FFN4 A0A195ED00
E0AE01 A0A212EXH0 A0A194QEU0 Q179Z8 A0A1S4GX24 A0A2J7Q1Z0 A0A084VUJ5 B0WEC7 A0A2J7Q1Y3 A0A182JDB4 A0A182IEK3 Q7Q3E3 A0A182X1M3 A0A182L2E6 A0A182TQV1 A0A182UTW7 A0A2J7Q1X9 A0A182F2Z9 A0A182QR49 A0A2J7Q1X8 A0A182JZU0 A0A182R3C0 A0A0A9WUN8 A0A182MFB7 A0A1Q3FYV3 A0A182GRF1 A0A182MZF5 A0A182WA31 A0A182SAY4 A0A0A9X221 A0A1B6CB83 A0A2P8YHA5 A0A1W4XUI1 A0A146M315 D6W6B9 A0A1B6E8U1 A0A131YVU5 A0A1B6CT37 N6UD82 E0VGF8 A0A023GCL6 A0A1W4UTJ4 A0A0J9R1G1 A0A182YGR6 Q8SY07 A0A0P9A5N5 B3N3D2 A0A1B6D349 A0A0R1DMD6 A0A1Y1N2C8 A0A1L8DJE2 A0A1L8DM79 A0A2R5LDK8 A0A1W4XTL2 V9ICV7 A0A293MTF9 A0A1W4XJH4 A0A0R3NTZ0 A0A131XNL6 A0A1B6CG67 A0A1V9XUE6 A0A0Q9X3U2 K7IYN3 A0A1B6D7R3 A0A0C9RVG5 A0A0A1WVU7 A0A3S3NWF1 W8C1N0 A0A147BTM3 A0A034VQN1 B4KIN4 A0A0K8V8K8 A0A0L7QNV3 A0A1W4V5W7 I6YMZ0 B3MNJ3 B4Q486 A0A0J9TM68 X2JDZ1 A0A0P4XQH1 B4MDL9 A0A0P4XJ46 A0A0M5J3W9 A0A0J9R150 A0A3S0ZXP0 A0A0Q5WJK4 V4AYC5 A0A2C9JFN1 A0A0M8ZNU5 A0A026WK07 T1FPB1 A0A0L8FFN4 A0A195ED00
Pubmed
19121390
21122821
22118469
26354079
17510324
12364791
+ More
24438588 20966253 25401762 26483478 26823975 29403074 18362917 19820115 26830274 23537049 20566863 22936249 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 28004739 15632085 28049606 28327890 20075255 25830018 24495485 29652888 25348373 22772333 23254933 15562597 24508170
24438588 20966253 25401762 26483478 26823975 29403074 18362917 19820115 26830274 23537049 20566863 22936249 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 28004739 15632085 28049606 28327890 20075255 25830018 24495485 29652888 25348373 22772333 23254933 15562597 24508170
EMBL
BABH01025562
ODYU01005656
SOQ46727.1
GDQN01000802
JAT90252.1
NWSH01000420
+ More
PCG76521.1 PCG76523.1 GDQN01000688 JAT90366.1 HM747956 ADK97802.1 AGBW02011772 OWR46161.1 KQ459054 KPJ03939.1 CH477342 EAT43073.1 AAAB01008964 NEVH01019376 PNF22593.1 ATLV01016849 ATLV01016850 KE525120 KFB41639.1 DS231907 EDS45487.1 PNF22594.1 APCN01005204 EAA12249.4 PNF22591.1 AXCN02000606 PNF22595.1 GBHO01032493 JAG11111.1 AXCM01010786 GFDL01002332 JAV32713.1 JXUM01016215 KQ560452 KXJ82350.1 GBHO01032494 GBRD01003810 GDHC01015827 JAG11110.1 JAG62011.1 JAQ02802.1 GEDC01026604 JAS10694.1 PYGN01000596 PSN43631.1 GDHC01005247 JAQ13382.1 KQ971319 EFA11378.1 GEDC01003024 JAS34274.1 GEDV01005912 JAP82645.1 GEDC01020803 JAS16495.1 APGK01033018 APGK01033019 KB740860 KB632042 ENN78571.1 ERL88238.1 DS235142 EEB12464.1 GBBM01003809 JAC31609.1 CM002910 KMY89951.1 AE014134 AY075464 AAF53213.2 AAL68277.1 CH902620 KPU73785.1 CH954177 EDV58772.2 GEDC01017177 JAS20121.1 CM000157 KRJ97645.1 GEZM01014752 JAV92012.1 GFDF01007597 JAV06487.1 GFDF01006644 JAV07440.1 GGLE01003422 MBY07548.1 JR039953 AEY58920.1 GFWV01018661 MAA43389.1 CH379061 KRT04578.1 GEFH01000833 JAP67748.1 GEDC01024919 GEDC01004871 JAS12379.1 JAS32427.1 MNPL01003910 OQR77127.1 CH933807 KRG02671.1 GEDC01015577 JAS21721.1 GBYB01012910 JAG82677.1 GBXI01011103 JAD03189.1 NCKU01002090 RWS10450.1 GAMC01010981 JAB95574.1 GEGO01001303 JAR94101.1 GAKP01015079 GAKP01015076 JAC43873.1 EDW11377.1 GDHF01017103 JAI35211.1 KQ414843 KOC60303.1 JX049540 AFN65700.1 EDV32101.1 CM000361 EDX04836.1 KMY89950.1 AHN54395.1 GDIP01240549 GDIP01053320 JAI82852.1 CH940661 EDW71280.1 GDIP01240550 GDIP01240548 JAI82851.1 CP012523 ALC39444.1 KMY89952.1 RQTK01000080 RUS88536.1 KQS70510.1 KB201234 ESO98636.1 KQ435955 KOX68045.1 KK107167 EZA56350.1 AMQM01007939 KB097694 ESN91862.1 KQ433417 KOF62442.1 KQ979074 KYN22719.1
PCG76521.1 PCG76523.1 GDQN01000688 JAT90366.1 HM747956 ADK97802.1 AGBW02011772 OWR46161.1 KQ459054 KPJ03939.1 CH477342 EAT43073.1 AAAB01008964 NEVH01019376 PNF22593.1 ATLV01016849 ATLV01016850 KE525120 KFB41639.1 DS231907 EDS45487.1 PNF22594.1 APCN01005204 EAA12249.4 PNF22591.1 AXCN02000606 PNF22595.1 GBHO01032493 JAG11111.1 AXCM01010786 GFDL01002332 JAV32713.1 JXUM01016215 KQ560452 KXJ82350.1 GBHO01032494 GBRD01003810 GDHC01015827 JAG11110.1 JAG62011.1 JAQ02802.1 GEDC01026604 JAS10694.1 PYGN01000596 PSN43631.1 GDHC01005247 JAQ13382.1 KQ971319 EFA11378.1 GEDC01003024 JAS34274.1 GEDV01005912 JAP82645.1 GEDC01020803 JAS16495.1 APGK01033018 APGK01033019 KB740860 KB632042 ENN78571.1 ERL88238.1 DS235142 EEB12464.1 GBBM01003809 JAC31609.1 CM002910 KMY89951.1 AE014134 AY075464 AAF53213.2 AAL68277.1 CH902620 KPU73785.1 CH954177 EDV58772.2 GEDC01017177 JAS20121.1 CM000157 KRJ97645.1 GEZM01014752 JAV92012.1 GFDF01007597 JAV06487.1 GFDF01006644 JAV07440.1 GGLE01003422 MBY07548.1 JR039953 AEY58920.1 GFWV01018661 MAA43389.1 CH379061 KRT04578.1 GEFH01000833 JAP67748.1 GEDC01024919 GEDC01004871 JAS12379.1 JAS32427.1 MNPL01003910 OQR77127.1 CH933807 KRG02671.1 GEDC01015577 JAS21721.1 GBYB01012910 JAG82677.1 GBXI01011103 JAD03189.1 NCKU01002090 RWS10450.1 GAMC01010981 JAB95574.1 GEGO01001303 JAR94101.1 GAKP01015079 GAKP01015076 JAC43873.1 EDW11377.1 GDHF01017103 JAI35211.1 KQ414843 KOC60303.1 JX049540 AFN65700.1 EDV32101.1 CM000361 EDX04836.1 KMY89950.1 AHN54395.1 GDIP01240549 GDIP01053320 JAI82852.1 CH940661 EDW71280.1 GDIP01240550 GDIP01240548 JAI82851.1 CP012523 ALC39444.1 KMY89952.1 RQTK01000080 RUS88536.1 KQS70510.1 KB201234 ESO98636.1 KQ435955 KOX68045.1 KK107167 EZA56350.1 AMQM01007939 KB097694 ESN91862.1 KQ433417 KOF62442.1 KQ979074 KYN22719.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000008820
UP000235965
+ More
UP000030765 UP000002320 UP000075880 UP000075840 UP000007062 UP000076407 UP000075882 UP000075902 UP000075903 UP000069272 UP000075886 UP000075881 UP000075900 UP000075883 UP000069940 UP000249989 UP000075884 UP000075920 UP000075901 UP000245037 UP000192223 UP000007266 UP000019118 UP000030742 UP000009046 UP000192221 UP000076408 UP000000803 UP000007801 UP000008711 UP000002282 UP000001819 UP000192247 UP000009192 UP000002358 UP000285301 UP000053825 UP000000304 UP000008792 UP000092553 UP000271974 UP000030746 UP000076420 UP000053105 UP000053097 UP000015101 UP000053454 UP000078492
UP000030765 UP000002320 UP000075880 UP000075840 UP000007062 UP000076407 UP000075882 UP000075902 UP000075903 UP000069272 UP000075886 UP000075881 UP000075900 UP000075883 UP000069940 UP000249989 UP000075884 UP000075920 UP000075901 UP000245037 UP000192223 UP000007266 UP000019118 UP000030742 UP000009046 UP000192221 UP000076408 UP000000803 UP000007801 UP000008711 UP000002282 UP000001819 UP000192247 UP000009192 UP000002358 UP000285301 UP000053825 UP000000304 UP000008792 UP000092553 UP000271974 UP000030746 UP000076420 UP000053105 UP000053097 UP000015101 UP000053454 UP000078492
Interpro
Gene 3D
ProteinModelPortal
H9JW29
A0A2H1W120
A0A1E1WTG6
A0A2A4JWZ9
A0A2A4JYG0
A0A1E1WU61
+ More
E0AE01 A0A212EXH0 A0A194QEU0 Q179Z8 A0A1S4GX24 A0A2J7Q1Z0 A0A084VUJ5 B0WEC7 A0A2J7Q1Y3 A0A182JDB4 A0A182IEK3 Q7Q3E3 A0A182X1M3 A0A182L2E6 A0A182TQV1 A0A182UTW7 A0A2J7Q1X9 A0A182F2Z9 A0A182QR49 A0A2J7Q1X8 A0A182JZU0 A0A182R3C0 A0A0A9WUN8 A0A182MFB7 A0A1Q3FYV3 A0A182GRF1 A0A182MZF5 A0A182WA31 A0A182SAY4 A0A0A9X221 A0A1B6CB83 A0A2P8YHA5 A0A1W4XUI1 A0A146M315 D6W6B9 A0A1B6E8U1 A0A131YVU5 A0A1B6CT37 N6UD82 E0VGF8 A0A023GCL6 A0A1W4UTJ4 A0A0J9R1G1 A0A182YGR6 Q8SY07 A0A0P9A5N5 B3N3D2 A0A1B6D349 A0A0R1DMD6 A0A1Y1N2C8 A0A1L8DJE2 A0A1L8DM79 A0A2R5LDK8 A0A1W4XTL2 V9ICV7 A0A293MTF9 A0A1W4XJH4 A0A0R3NTZ0 A0A131XNL6 A0A1B6CG67 A0A1V9XUE6 A0A0Q9X3U2 K7IYN3 A0A1B6D7R3 A0A0C9RVG5 A0A0A1WVU7 A0A3S3NWF1 W8C1N0 A0A147BTM3 A0A034VQN1 B4KIN4 A0A0K8V8K8 A0A0L7QNV3 A0A1W4V5W7 I6YMZ0 B3MNJ3 B4Q486 A0A0J9TM68 X2JDZ1 A0A0P4XQH1 B4MDL9 A0A0P4XJ46 A0A0M5J3W9 A0A0J9R150 A0A3S0ZXP0 A0A0Q5WJK4 V4AYC5 A0A2C9JFN1 A0A0M8ZNU5 A0A026WK07 T1FPB1 A0A0L8FFN4 A0A195ED00
E0AE01 A0A212EXH0 A0A194QEU0 Q179Z8 A0A1S4GX24 A0A2J7Q1Z0 A0A084VUJ5 B0WEC7 A0A2J7Q1Y3 A0A182JDB4 A0A182IEK3 Q7Q3E3 A0A182X1M3 A0A182L2E6 A0A182TQV1 A0A182UTW7 A0A2J7Q1X9 A0A182F2Z9 A0A182QR49 A0A2J7Q1X8 A0A182JZU0 A0A182R3C0 A0A0A9WUN8 A0A182MFB7 A0A1Q3FYV3 A0A182GRF1 A0A182MZF5 A0A182WA31 A0A182SAY4 A0A0A9X221 A0A1B6CB83 A0A2P8YHA5 A0A1W4XUI1 A0A146M315 D6W6B9 A0A1B6E8U1 A0A131YVU5 A0A1B6CT37 N6UD82 E0VGF8 A0A023GCL6 A0A1W4UTJ4 A0A0J9R1G1 A0A182YGR6 Q8SY07 A0A0P9A5N5 B3N3D2 A0A1B6D349 A0A0R1DMD6 A0A1Y1N2C8 A0A1L8DJE2 A0A1L8DM79 A0A2R5LDK8 A0A1W4XTL2 V9ICV7 A0A293MTF9 A0A1W4XJH4 A0A0R3NTZ0 A0A131XNL6 A0A1B6CG67 A0A1V9XUE6 A0A0Q9X3U2 K7IYN3 A0A1B6D7R3 A0A0C9RVG5 A0A0A1WVU7 A0A3S3NWF1 W8C1N0 A0A147BTM3 A0A034VQN1 B4KIN4 A0A0K8V8K8 A0A0L7QNV3 A0A1W4V5W7 I6YMZ0 B3MNJ3 B4Q486 A0A0J9TM68 X2JDZ1 A0A0P4XQH1 B4MDL9 A0A0P4XJ46 A0A0M5J3W9 A0A0J9R150 A0A3S0ZXP0 A0A0Q5WJK4 V4AYC5 A0A2C9JFN1 A0A0M8ZNU5 A0A026WK07 T1FPB1 A0A0L8FFN4 A0A195ED00
PDB
3HPK
E-value=1.80084e-37,
Score=392
Ontologies
GO
GO:0019904
GO:0016301
GO:0002092
GO:0014069
GO:0005543
GO:0005886
GO:0043005
GO:0008021
GO:0043113
GO:0006886
GO:0045202
GO:0005737
GO:0097062
GO:0097061
GO:0032588
GO:0005080
GO:0098842
GO:0048639
GO:0071329
GO:0045055
GO:0043025
GO:0099175
GO:0005515
GO:0005840
GO:0006412
GO:0033557
GO:0005839
GO:0051603
GO:0046835
GO:0006281
GO:0006259
PANTHER
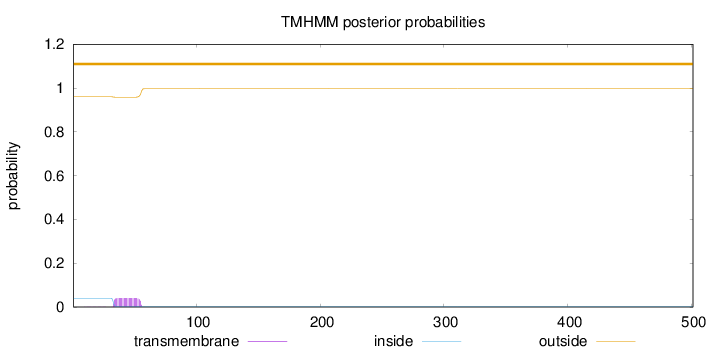
Topology
Length:
501
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.9031
Exp number, first 60 AAs:
0.90029
Total prob of N-in:
0.03968
outside
1 - 501
Population Genetic Test Statistics
Pi
296.423712
Theta
168.190172
Tajima's D
2.874543
CLR
0.236982
CSRT
0.972101394930253
Interpretation
Uncertain
