Gene
KWMTBOMO16375 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013801
Annotation
PREDICTED:_charged_multivesicular_body_protein_4b_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.902 Nuclear Reliability : 2.204
Sequence
CDS
ATGAGTTTTCTCGGGAAAATATTCGGTGGCAAAAAAGAAGAGAAGGGGCCCACTACGCATGAAGCTATCCAAAAACTCCGTGAAACTGAAGAATTGCTGATCAAGAAGCAAGAGTTTTTGGAAAAGAAGATCGATTTAGAAGTACAGACAGCCAGGAAACATGGCACTAAGAACAAAAGAGCTGCCATTGCAGCTCTCAAGCGGAAGAAGAGATACGAGAAGCAACTTACTCAAATAGATGGGACCCTCACTCAGATTGAGGCCCAAAGGGAAGCGCTAGAAGGTGCCAATACCAATGCCCAAGTATTGAACACTATGCGCGAGGCAGCTAATGCCATGAAGCTCGCACACAAAGACATTGATGTGGACAAAGTACACGATATCATGGATGATATTGCTGAACAACACGACATCTCTCGTGAGATCACCGAGGCGATCAGTAACAATGTGGCCTTCCCTAATGATATAGATGAAGATGAACTCGAGAAGGAACTTGAAGAATTAGAACAGGAGGATCTGGACAAGGAGATGCTGGGCATCAACGTGCCGACGGACACGCTGCCCGATGTCCCGGCCTCGGAACTGGTACACGACAAACCCAAACCGAGCAAGTCGAAACAGACAGAGGATGACGATGAGCTTGCTCAGCTGCAGTCGTGGGCCACATAG
Protein
MSFLGKIFGGKKEEKGPTTHEAIQKLRETEELLIKKQEFLEKKIDLEVQTARKHGTKNKRAAIAALKRKKRYEKQLTQIDGTLTQIEAQREALEGANTNAQVLNTMREAANAMKLAHKDIDVDKVHDIMDDIAEQHDISREITEAISNNVAFPNDIDEDELEKELEELEQEDLDKEMLGINVPTDTLPDVPASELVHDKPKPSKSKQTEDDDELAQLQSWAT
Summary
Uniprot
A0A2A4JXJ1
A0A1V0JHU9
A0A194QKF6
H9JW87
A0A0N1IJ71
S4NSU7
+ More
A0A3S2LCE9 A0A212ETP8 A0A0N1PGQ6 A0A1B6IE32 A0A0L7RCT8 A0A1W4WVV1 A0A1B6LBV6 A0A067R236 A0A0J7KW61 A0A0C9PIQ5 A0A151JP72 F4WBC6 A0A151JZM8 A0A158NAX0 A0A151X9R1 R4G4L2 A0A0P4X966 A0A151IP09 A0A1D2NG42 A0A232FE85 A0A1S3CXS3 A0A0P5FIT0 A0A0T6B0F6 E0VAJ6 A0A023F9J3 E2C316 A0A0V0G653 A0A1B6H014 K7IP52 A0A0A9Y694 D1FQ45 A0A0P5X495 A0A0P5TD42 A0A0M8ZZM3 A0A088ARB1 E9HGK4 A0A2R7WAE3 V9IKA6 A0A1Y1LPA9 A0A2A3EDI6 A0A084W2H9 A0A023EMD8 A0A0A1XJD6 W8BU09 A0A226DMN1 Q17KG9 A0A0K8VN15 E2A5M9 A0A034WDQ3 A0A0M5J4V0 A0A182MN93 B4NNL3 A0A151HY47 A0A026WRU2 A0A0N7ZGD6 A0A0K8TT00 A0A182TP28 A0A2M4AB03 A0A2M4AAZ0 A0A182N880 A0A182Q9X8 A0A3F2YSS7 A0A182VBT5 A0A182XD53 A0A182KVZ3 Q7Q7G8 W5JRM1 A0A2M4CX45 A0A182TBV8 B3MDH4 B4ME56 A0A2M3Z3J7 A0A0B2VM91 D3TLF1 A0A182IRJ4 A0A1A9ZNQ5 A0A1A9VY33 A0A085NN14 A0A0N5D2H2 A0A1L8EE00 A0A154PIW9 B4KQL4 A0A077Z7D9 A0A1B0B239 A0A1A9WMU2 A0A1A9XJA4 T1E8Y5 F1L3E1 A0A0Q9WW21 A0A183I754 A0A1L8EMW7 A0A3R7PXM5 A0A0H5SMH3
A0A3S2LCE9 A0A212ETP8 A0A0N1PGQ6 A0A1B6IE32 A0A0L7RCT8 A0A1W4WVV1 A0A1B6LBV6 A0A067R236 A0A0J7KW61 A0A0C9PIQ5 A0A151JP72 F4WBC6 A0A151JZM8 A0A158NAX0 A0A151X9R1 R4G4L2 A0A0P4X966 A0A151IP09 A0A1D2NG42 A0A232FE85 A0A1S3CXS3 A0A0P5FIT0 A0A0T6B0F6 E0VAJ6 A0A023F9J3 E2C316 A0A0V0G653 A0A1B6H014 K7IP52 A0A0A9Y694 D1FQ45 A0A0P5X495 A0A0P5TD42 A0A0M8ZZM3 A0A088ARB1 E9HGK4 A0A2R7WAE3 V9IKA6 A0A1Y1LPA9 A0A2A3EDI6 A0A084W2H9 A0A023EMD8 A0A0A1XJD6 W8BU09 A0A226DMN1 Q17KG9 A0A0K8VN15 E2A5M9 A0A034WDQ3 A0A0M5J4V0 A0A182MN93 B4NNL3 A0A151HY47 A0A026WRU2 A0A0N7ZGD6 A0A0K8TT00 A0A182TP28 A0A2M4AB03 A0A2M4AAZ0 A0A182N880 A0A182Q9X8 A0A3F2YSS7 A0A182VBT5 A0A182XD53 A0A182KVZ3 Q7Q7G8 W5JRM1 A0A2M4CX45 A0A182TBV8 B3MDH4 B4ME56 A0A2M3Z3J7 A0A0B2VM91 D3TLF1 A0A182IRJ4 A0A1A9ZNQ5 A0A1A9VY33 A0A085NN14 A0A0N5D2H2 A0A1L8EE00 A0A154PIW9 B4KQL4 A0A077Z7D9 A0A1B0B239 A0A1A9WMU2 A0A1A9XJA4 T1E8Y5 F1L3E1 A0A0Q9WW21 A0A183I754 A0A1L8EMW7 A0A3R7PXM5 A0A0H5SMH3
Pubmed
26354079
19121390
23622113
22118469
24845553
21719571
+ More
21347285 27289101 28648823 20566863 25474469 20798317 20075255 25401762 26823975 21292972 28004739 24438588 24945155 26483478 25830018 24495485 17510324 25348373 17994087 24508170 30249741 26369729 20966253 12364791 14747013 17210077 20920257 23761445 20353571 24929829 21685128 27762356 17885136
21347285 27289101 28648823 20566863 25474469 20798317 20075255 25401762 26823975 21292972 28004739 24438588 24945155 26483478 25830018 24495485 17510324 25348373 17994087 24508170 30249741 26369729 20966253 12364791 14747013 17210077 20920257 23761445 20353571 24929829 21685128 27762356 17885136
EMBL
NWSH01000420
PCG76519.1
KY694526
ARD08867.1
KQ459054
KPJ03941.1
+ More
BABH01025560 KQ459999 KPJ18492.1 GAIX01012426 JAA80134.1 RSAL01000240 RVE43680.1 AGBW02012535 OWR44875.1 KQ458517 KPJ05848.1 GECU01022556 JAS85150.1 KQ414615 KOC68605.1 GEBQ01018809 JAT21168.1 KK852981 KDR12954.1 LBMM01002492 KMQ94747.1 GBYB01000828 JAG70595.1 KQ978756 KYN28583.1 GL888062 EGI68482.1 KQ981451 KYN41606.1 ADTU01010426 ADTU01010427 ADTU01010428 KQ982365 KYQ57103.1 ACPB03002868 GAHY01000676 JAA76834.1 GDIP01246412 JAI76989.1 KQ976899 KYN07203.1 LJIJ01000050 ODN04233.1 NNAY01000372 OXU28820.1 GDIQ01256529 JAJ95195.1 LJIG01016324 KRT80943.1 DS235006 EEB10402.1 GBBI01000560 JAC18152.1 GL452257 EFN77673.1 GECL01002661 JAP03463.1 GECZ01001764 JAS68005.1 GBHO01015897 GBHO01015895 GBRD01001571 GDHC01019754 JAG27707.1 JAG27709.1 JAG64250.1 JAP98874.1 EZ419945 ACZ28300.1 GDIP01077912 JAM25803.1 GDIP01129160 JAL74554.1 KQ435791 KOX74360.1 GL732642 EFX69136.1 KK854513 PTY16488.1 JR051415 AEY61495.1 GEZM01051861 JAV74658.1 KZ288289 PBC29289.1 ATLV01019619 KE525275 KFB44423.1 JXUM01008014 JXUM01008015 GAPW01004119 KQ560251 JAC09479.1 KXJ83558.1 GBXI01003230 GBXI01000184 JAD11062.1 JAD14108.1 GAMC01003873 GAMC01003872 JAC02683.1 LNIX01000017 OXA45476.1 CH477224 EAT47182.1 GDHF01012042 JAI40272.1 GL437023 EFN71226.1 GAKP01007029 JAC51923.1 CP012524 ALC41568.1 AXCM01004465 AXCM01012800 CH964282 EDW85952.1 KQ976728 KYM76497.1 KK107119 QOIP01000009 EZA58770.1 RLU18856.1 GDIP01248049 JAI75352.1 GDAI01000335 JAI17268.1 GGFK01004643 MBW37964.1 GGFK01004624 MBW37945.1 AXCN02000959 APCN01004443 AAAB01008960 EAA11799.3 ADMH02000550 ETN65923.1 GGFL01005661 MBW69839.1 CH902619 EDV36422.1 CH940662 EDW58821.1 GGFM01002324 MBW23075.1 JPKZ01001349 UYWY01021622 KHN82474.1 VDM44551.1 CCAG010007048 EZ422253 ADD18529.1 KL363186 KL367485 KFD57958.1 KFD70860.1 UYYF01004472 VDN04495.1 GFDG01001980 JAV16819.1 KQ434931 KZC11767.1 CH933808 EDW08183.1 HG805970 CDW55664.1 JXJN01007396 GAMD01002348 JAA99242.1 JI170585 ADY44645.1 KRG00319.1 UZAJ01042378 VDP22615.1 CM004483 OCT60619.1 QCYY01001130 ROT80330.1 LN856992 CRZ24917.1
BABH01025560 KQ459999 KPJ18492.1 GAIX01012426 JAA80134.1 RSAL01000240 RVE43680.1 AGBW02012535 OWR44875.1 KQ458517 KPJ05848.1 GECU01022556 JAS85150.1 KQ414615 KOC68605.1 GEBQ01018809 JAT21168.1 KK852981 KDR12954.1 LBMM01002492 KMQ94747.1 GBYB01000828 JAG70595.1 KQ978756 KYN28583.1 GL888062 EGI68482.1 KQ981451 KYN41606.1 ADTU01010426 ADTU01010427 ADTU01010428 KQ982365 KYQ57103.1 ACPB03002868 GAHY01000676 JAA76834.1 GDIP01246412 JAI76989.1 KQ976899 KYN07203.1 LJIJ01000050 ODN04233.1 NNAY01000372 OXU28820.1 GDIQ01256529 JAJ95195.1 LJIG01016324 KRT80943.1 DS235006 EEB10402.1 GBBI01000560 JAC18152.1 GL452257 EFN77673.1 GECL01002661 JAP03463.1 GECZ01001764 JAS68005.1 GBHO01015897 GBHO01015895 GBRD01001571 GDHC01019754 JAG27707.1 JAG27709.1 JAG64250.1 JAP98874.1 EZ419945 ACZ28300.1 GDIP01077912 JAM25803.1 GDIP01129160 JAL74554.1 KQ435791 KOX74360.1 GL732642 EFX69136.1 KK854513 PTY16488.1 JR051415 AEY61495.1 GEZM01051861 JAV74658.1 KZ288289 PBC29289.1 ATLV01019619 KE525275 KFB44423.1 JXUM01008014 JXUM01008015 GAPW01004119 KQ560251 JAC09479.1 KXJ83558.1 GBXI01003230 GBXI01000184 JAD11062.1 JAD14108.1 GAMC01003873 GAMC01003872 JAC02683.1 LNIX01000017 OXA45476.1 CH477224 EAT47182.1 GDHF01012042 JAI40272.1 GL437023 EFN71226.1 GAKP01007029 JAC51923.1 CP012524 ALC41568.1 AXCM01004465 AXCM01012800 CH964282 EDW85952.1 KQ976728 KYM76497.1 KK107119 QOIP01000009 EZA58770.1 RLU18856.1 GDIP01248049 JAI75352.1 GDAI01000335 JAI17268.1 GGFK01004643 MBW37964.1 GGFK01004624 MBW37945.1 AXCN02000959 APCN01004443 AAAB01008960 EAA11799.3 ADMH02000550 ETN65923.1 GGFL01005661 MBW69839.1 CH902619 EDV36422.1 CH940662 EDW58821.1 GGFM01002324 MBW23075.1 JPKZ01001349 UYWY01021622 KHN82474.1 VDM44551.1 CCAG010007048 EZ422253 ADD18529.1 KL363186 KL367485 KFD57958.1 KFD70860.1 UYYF01004472 VDN04495.1 GFDG01001980 JAV16819.1 KQ434931 KZC11767.1 CH933808 EDW08183.1 HG805970 CDW55664.1 JXJN01007396 GAMD01002348 JAA99242.1 JI170585 ADY44645.1 KRG00319.1 UZAJ01042378 VDP22615.1 CM004483 OCT60619.1 QCYY01001130 ROT80330.1 LN856992 CRZ24917.1
Proteomes
UP000218220
UP000053268
UP000005204
UP000053240
UP000283053
UP000007151
+ More
UP000053825 UP000192223 UP000027135 UP000036403 UP000078492 UP000007755 UP000078541 UP000005205 UP000075809 UP000015103 UP000078542 UP000094527 UP000215335 UP000079169 UP000009046 UP000008237 UP000002358 UP000053105 UP000005203 UP000000305 UP000242457 UP000030765 UP000069940 UP000249989 UP000198287 UP000008820 UP000000311 UP000092553 UP000075883 UP000007798 UP000078540 UP000053097 UP000279307 UP000075902 UP000075884 UP000075886 UP000075840 UP000075903 UP000076407 UP000075882 UP000007062 UP000000673 UP000075901 UP000007801 UP000008792 UP000031036 UP000050794 UP000267007 UP000092444 UP000075880 UP000092445 UP000078200 UP000046394 UP000276776 UP000076502 UP000009192 UP000030665 UP000092460 UP000091820 UP000092443 UP000050787 UP000186698 UP000283509
UP000053825 UP000192223 UP000027135 UP000036403 UP000078492 UP000007755 UP000078541 UP000005205 UP000075809 UP000015103 UP000078542 UP000094527 UP000215335 UP000079169 UP000009046 UP000008237 UP000002358 UP000053105 UP000005203 UP000000305 UP000242457 UP000030765 UP000069940 UP000249989 UP000198287 UP000008820 UP000000311 UP000092553 UP000075883 UP000007798 UP000078540 UP000053097 UP000279307 UP000075902 UP000075884 UP000075886 UP000075840 UP000075903 UP000076407 UP000075882 UP000007062 UP000000673 UP000075901 UP000007801 UP000008792 UP000031036 UP000050794 UP000267007 UP000092444 UP000075880 UP000092445 UP000078200 UP000046394 UP000276776 UP000076502 UP000009192 UP000030665 UP000092460 UP000091820 UP000092443 UP000050787 UP000186698 UP000283509
PRIDE
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2A4JXJ1
A0A1V0JHU9
A0A194QKF6
H9JW87
A0A0N1IJ71
S4NSU7
+ More
A0A3S2LCE9 A0A212ETP8 A0A0N1PGQ6 A0A1B6IE32 A0A0L7RCT8 A0A1W4WVV1 A0A1B6LBV6 A0A067R236 A0A0J7KW61 A0A0C9PIQ5 A0A151JP72 F4WBC6 A0A151JZM8 A0A158NAX0 A0A151X9R1 R4G4L2 A0A0P4X966 A0A151IP09 A0A1D2NG42 A0A232FE85 A0A1S3CXS3 A0A0P5FIT0 A0A0T6B0F6 E0VAJ6 A0A023F9J3 E2C316 A0A0V0G653 A0A1B6H014 K7IP52 A0A0A9Y694 D1FQ45 A0A0P5X495 A0A0P5TD42 A0A0M8ZZM3 A0A088ARB1 E9HGK4 A0A2R7WAE3 V9IKA6 A0A1Y1LPA9 A0A2A3EDI6 A0A084W2H9 A0A023EMD8 A0A0A1XJD6 W8BU09 A0A226DMN1 Q17KG9 A0A0K8VN15 E2A5M9 A0A034WDQ3 A0A0M5J4V0 A0A182MN93 B4NNL3 A0A151HY47 A0A026WRU2 A0A0N7ZGD6 A0A0K8TT00 A0A182TP28 A0A2M4AB03 A0A2M4AAZ0 A0A182N880 A0A182Q9X8 A0A3F2YSS7 A0A182VBT5 A0A182XD53 A0A182KVZ3 Q7Q7G8 W5JRM1 A0A2M4CX45 A0A182TBV8 B3MDH4 B4ME56 A0A2M3Z3J7 A0A0B2VM91 D3TLF1 A0A182IRJ4 A0A1A9ZNQ5 A0A1A9VY33 A0A085NN14 A0A0N5D2H2 A0A1L8EE00 A0A154PIW9 B4KQL4 A0A077Z7D9 A0A1B0B239 A0A1A9WMU2 A0A1A9XJA4 T1E8Y5 F1L3E1 A0A0Q9WW21 A0A183I754 A0A1L8EMW7 A0A3R7PXM5 A0A0H5SMH3
A0A3S2LCE9 A0A212ETP8 A0A0N1PGQ6 A0A1B6IE32 A0A0L7RCT8 A0A1W4WVV1 A0A1B6LBV6 A0A067R236 A0A0J7KW61 A0A0C9PIQ5 A0A151JP72 F4WBC6 A0A151JZM8 A0A158NAX0 A0A151X9R1 R4G4L2 A0A0P4X966 A0A151IP09 A0A1D2NG42 A0A232FE85 A0A1S3CXS3 A0A0P5FIT0 A0A0T6B0F6 E0VAJ6 A0A023F9J3 E2C316 A0A0V0G653 A0A1B6H014 K7IP52 A0A0A9Y694 D1FQ45 A0A0P5X495 A0A0P5TD42 A0A0M8ZZM3 A0A088ARB1 E9HGK4 A0A2R7WAE3 V9IKA6 A0A1Y1LPA9 A0A2A3EDI6 A0A084W2H9 A0A023EMD8 A0A0A1XJD6 W8BU09 A0A226DMN1 Q17KG9 A0A0K8VN15 E2A5M9 A0A034WDQ3 A0A0M5J4V0 A0A182MN93 B4NNL3 A0A151HY47 A0A026WRU2 A0A0N7ZGD6 A0A0K8TT00 A0A182TP28 A0A2M4AB03 A0A2M4AAZ0 A0A182N880 A0A182Q9X8 A0A3F2YSS7 A0A182VBT5 A0A182XD53 A0A182KVZ3 Q7Q7G8 W5JRM1 A0A2M4CX45 A0A182TBV8 B3MDH4 B4ME56 A0A2M3Z3J7 A0A0B2VM91 D3TLF1 A0A182IRJ4 A0A1A9ZNQ5 A0A1A9VY33 A0A085NN14 A0A0N5D2H2 A0A1L8EE00 A0A154PIW9 B4KQL4 A0A077Z7D9 A0A1B0B239 A0A1A9WMU2 A0A1A9XJA4 T1E8Y5 F1L3E1 A0A0Q9WW21 A0A183I754 A0A1L8EMW7 A0A3R7PXM5 A0A0H5SMH3
PDB
5J45
E-value=8.72207e-27,
Score=296
Ontologies
GO
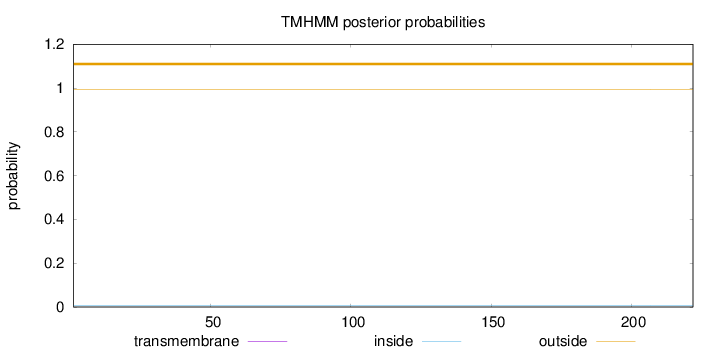
Topology
Length:
222
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00720
outside
1 - 222
Population Genetic Test Statistics
Pi
29.378917
Theta
22.952708
Tajima's D
2.357958
CLR
0.075305
CSRT
0.932453377331133
Interpretation
Uncertain
