Gene
KWMTBOMO16373
Pre Gene Modal
BGIBMGA013800
Annotation
Tektin-4_[Papilio_machaon]
Full name
Histone acetyltransferase
Location in the cell
Nuclear Reliability : 3.536
Sequence
CDS
ATGAATTCAGCCGAAAATGGAGATTTACTTTCGTATGGCGATAAGAACTATGGTCTCGGTGCGATGCAGAATTGGCATGAACACAACGAGCGAATCCTGAAAGGCGACATCATCAGAACAACTGACGATGAAACATCCATAACTAAAAATTGCATCATCAAAATCAACAATACATCTTGTAACGACTTCGCGGACACAACCGGCCAGTTGAAACAGAGAGCCAGACACATCAACTACTGGCAGGCCGAGCTAGAGAGAGCTATAAGGGACATTGACGCCGAATCAGACGTATTGGAAGAGCAGAGGCAACAGCTCAAAAACGCTGTGGACGTATTAAAGATTCCGGAATACGTGACGGAAGAATGTTTGGACGTTAGACGTAACAGAATGCAATCGGACCTCATCTACGATCCGCCTCAAGAGGAACTATTCACCGAATCTGCTCTCATTGAAAACGTTAAACGTTTGCACGGCGACATGATTAGAGACATAGAAAATCAGATGAAAGTTAATTTAGCAGCCAAACATTCTCTGGAGAGGGACTGGAGTAATAAGCACTTGGCGTTTAAATACGAAACCGCTAATTCAGAGTTAGAAACGAAATCGGTAAACGTGAAATATTCGCCAGGAGCCACCAGACTCTGCGAAGGACAATCCGACGTCCCATCTTGGGAGGGTCACACTGTTAACGCACTAGATCAGTTTAAAGCCGATCTAGATTCGTCACAGCAGCTCAGAAAGAGAGTTGAAGCTACCATATTGAACACAGCACGAGATTTAAGGAGCCAAGATCTGAAAGTGAACAGGGCGATCGCCGAGCGCATCGCCAGAACCCAACAAGTGAAAACCGAATTGGAAAACCAACTGAGATTGACGATACAAAAGATCGGCGAAACGGAAAATATTCTAGAGACGCTGCACGAGGAAACTATGAAGGTTGCCCAGAGGATCCAAGTCGCGCAGACGAGGTTGAACACAAAGAACAACAGACCAACAGTTGAGAATTGCAGAGAAGGTTCCCTTTTCAGTTTGATCGAAGAAGTCCGAGATCTGAACGACAGTATGAGTCTACTACGTAAACGATCTCTGGTAACGGAGAAATTGAGAGCGGAACTGATCCAGGAGAGGTCTCTATTGGAAAACGAAATAACAGTGAAGAATAAAACTATCCATATAGATGATGAACGATGCTTGTTTCTGAGGTCGCATTTTCAATCAGCTGAGAAATTGAGCGGTTTCTAA
Protein
MNSAENGDLLSYGDKNYGLGAMQNWHEHNERILKGDIIRTTDDETSITKNCIIKINNTSCNDFADTTGQLKQRARHINYWQAELERAIRDIDAESDVLEEQRQQLKNAVDVLKIPEYVTEECLDVRRNRMQSDLIYDPPQEELFTESALIENVKRLHGDMIRDIENQMKVNLAAKHSLERDWSNKHLAFKYETANSELETKSVNVKYSPGATRLCEGQSDVPSWEGHTVNALDQFKADLDSSQQLRKRVEATILNTARDLRSQDLKVNRAIAERIARTQQVKTELENQLRLTIQKIGETENILETLHEETMKVAQRIQVAQTRLNTKNNRPTVENCREGSLFSLIEEVRDLNDSMSLLRKRSLVTEKLRAELIQERSLLENEITVKNKTIHIDDERCLFLRSHFQSAEKLSGF
Summary
Catalytic Activity
acetyl-CoA + L-lysyl-[protein] = CoA + H(+) + N(6)-acetyl-L-lysyl-[protein]
Similarity
Belongs to the MYST (SAS/MOZ) family.
Uniprot
H9JW86
A0A0N1IBU9
A0A212F8N0
A0A194PJK5
A0A0L7LCK7
H9JWM1
+ More
A0A2A4K8G5 A0A1I8MQ14 A0A2W1BDL7 A0A2J7Q0W3 A0A194Q0S4 A0A1J1IHY3 A0A2A4KA34 A0A0L7LJZ3 A0A0N1IQ59 A0A1B6BY47 B0WGB8 A0A2W1BI80 A0A1B0CEK9 Q7Q108 A0A023ET07 A0A212FLC4 A0A182SIM5 Q171C6 A0A182W8M4 A0A182XHD2 A0A182JDH7 A0A034W300 A0A1I8JTJ3 A0A0K8V0M0 A0A182VFR4 A0A182F336 A0A182GU33 A0A182M6S8 A0A182PDW9 A0A084VD10 A0A1B6MD53 A0A1B6GTT4 A0A0A1X419 W5JV24 A0A336LID9 A0A0L0CED8 A0A182K1J2 A0A182NNY0 A0A182TIT4 A0A0K8UHJ6 A0A182R1T9 A0A1B0D6I8 A0A067QTU0 A0A1A9V1K0 A0A2H1WKT9 A0A182PEF9 A0A1A9WUB4 W8BRM4 A0A1A9Y8T0 B4N030 A0A1B0BQ23 Q29KE0 B4H9F2 B3ML13 A0A1B0FL25 U4UPD2 B4M8U9 Q9BLD7 A0A1A9ZWA8 A0A026W8B8 H9JWM2 A0A3B0K3C4 A0A182HDJ0 B4Q6I3 A0A1W4U4P9 A0A3L8DBQ9 B4JED5 B3N620 B4KE95 B4NY77 B3PA87 A0A0L7LKB4 A0A2W1BAS8 Q9V3M9 A0A0J9R2F1 B4I1X5 A0A0J7NDZ5 A0A0M5IZ51 A0A154P562 A0A2A4K9W1 A0A224XT22 D6WUN0 T1I5P5 A0A182Y659 A0A212FLB2 A0A0M4ES94 A0A2C9JZH2 A0A1I8FZ59 A0A267EN10 A0A151X2K1 A0A267EH03 H2YEY8 A0A2R7WMD4
A0A2A4K8G5 A0A1I8MQ14 A0A2W1BDL7 A0A2J7Q0W3 A0A194Q0S4 A0A1J1IHY3 A0A2A4KA34 A0A0L7LJZ3 A0A0N1IQ59 A0A1B6BY47 B0WGB8 A0A2W1BI80 A0A1B0CEK9 Q7Q108 A0A023ET07 A0A212FLC4 A0A182SIM5 Q171C6 A0A182W8M4 A0A182XHD2 A0A182JDH7 A0A034W300 A0A1I8JTJ3 A0A0K8V0M0 A0A182VFR4 A0A182F336 A0A182GU33 A0A182M6S8 A0A182PDW9 A0A084VD10 A0A1B6MD53 A0A1B6GTT4 A0A0A1X419 W5JV24 A0A336LID9 A0A0L0CED8 A0A182K1J2 A0A182NNY0 A0A182TIT4 A0A0K8UHJ6 A0A182R1T9 A0A1B0D6I8 A0A067QTU0 A0A1A9V1K0 A0A2H1WKT9 A0A182PEF9 A0A1A9WUB4 W8BRM4 A0A1A9Y8T0 B4N030 A0A1B0BQ23 Q29KE0 B4H9F2 B3ML13 A0A1B0FL25 U4UPD2 B4M8U9 Q9BLD7 A0A1A9ZWA8 A0A026W8B8 H9JWM2 A0A3B0K3C4 A0A182HDJ0 B4Q6I3 A0A1W4U4P9 A0A3L8DBQ9 B4JED5 B3N620 B4KE95 B4NY77 B3PA87 A0A0L7LKB4 A0A2W1BAS8 Q9V3M9 A0A0J9R2F1 B4I1X5 A0A0J7NDZ5 A0A0M5IZ51 A0A154P562 A0A2A4K9W1 A0A224XT22 D6WUN0 T1I5P5 A0A182Y659 A0A212FLB2 A0A0M4ES94 A0A2C9JZH2 A0A1I8FZ59 A0A267EN10 A0A151X2K1 A0A267EH03 H2YEY8 A0A2R7WMD4
EC Number
2.3.1.48
Pubmed
19121390
26354079
22118469
26227816
25315136
28756777
+ More
12364791 14747013 17210077 24945155 17510324 25348373 26483478 24438588 25830018 20920257 23761445 26108605 24845553 24495485 17994087 15632085 23537049 12431405 24508170 30249741 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 18362917 19820115 25244985 15562597 26392545
12364791 14747013 17210077 24945155 17510324 25348373 26483478 24438588 25830018 20920257 23761445 26108605 24845553 24495485 17994087 15632085 23537049 12431405 24508170 30249741 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 18362917 19820115 25244985 15562597 26392545
EMBL
BABH01025560
KQ459999
KPJ18494.1
AGBW02009696
OWR50094.1
KQ459602
+ More
KPI93218.1 JTDY01001770 KOB72936.1 BABH01018360 BABH01018361 NWSH01000031 PCG80511.1 KZ150184 PZC72471.1 NEVH01019960 PNF22228.1 KQ459583 KPI98594.1 CVRI01000054 CRK99861.1 PCG80512.1 JTDY01000784 KOB75888.1 KQ459764 KPJ20100.1 GEDC01031107 JAS06191.1 DS231924 EDS26856.1 PZC72470.1 AJWK01008967 AAAB01008980 EAA14054.4 GAPW01001166 JAC12432.1 AGBW02007806 OWR54532.1 CH477459 EAT40600.1 GAKP01008996 JAC49956.1 APCN01001644 GDHF01019860 JAI32454.1 JXUM01018539 JXUM01018540 JXUM01018541 JXUM01018542 KQ560515 KXJ82034.1 AXCM01008115 ATLV01011119 KE524642 KFB35854.1 GEBQ01006132 JAT33845.1 GECZ01003930 JAS65839.1 GBXI01016390 GBXI01008465 JAC97901.1 JAD05827.1 ADMH02000277 ETN67143.1 UFQT01000011 SSX17590.1 JRES01000498 KNC30773.1 GDHF01026519 JAI25795.1 AJVK01003621 KK852945 KDR13451.1 ODYU01009337 SOQ53691.1 GAMC01002610 JAC03946.1 CH963920 EDW77965.1 JXJN01018354 CH379061 EAL33235.2 CH479228 EDW35384.1 CH902620 EDV30671.1 CCAG010020516 KB632308 ERL92021.1 CH940654 EDW57625.1 AB056651 BAB33388.1 KK107348 EZA52208.1 OUUW01000006 SPP82440.1 JXUM01034205 JXUM01034206 KQ561016 KXJ80024.1 CM000361 EDX05165.1 QOIP01000010 RLU17935.1 CH916368 EDW03655.1 CH954177 EDV58058.1 CH933807 EDW11840.1 CM000157 EDW89713.1 CH954297 EDV47705.1 KOB75887.1 PZC72472.1 AE014134 AY058398 AAF53488.1 AAL13627.1 AGB93011.1 CM002910 KMY90417.1 CH480820 EDW54532.1 LBMM01006278 KMQ90745.1 CP012523 ALC39886.1 KQ434820 KZC07027.1 PCG80510.1 GFTR01005315 JAW11111.1 KQ971357 EFA08354.1 ACPB03003574 OWR54533.1 ALC39895.1 NIVC01002675 NIVC01001955 PAA56021.1 PAA62277.1 KQ982578 KYQ54626.1 NIVC01002189 PAA60139.1 KK854992 PTY20160.1
KPI93218.1 JTDY01001770 KOB72936.1 BABH01018360 BABH01018361 NWSH01000031 PCG80511.1 KZ150184 PZC72471.1 NEVH01019960 PNF22228.1 KQ459583 KPI98594.1 CVRI01000054 CRK99861.1 PCG80512.1 JTDY01000784 KOB75888.1 KQ459764 KPJ20100.1 GEDC01031107 JAS06191.1 DS231924 EDS26856.1 PZC72470.1 AJWK01008967 AAAB01008980 EAA14054.4 GAPW01001166 JAC12432.1 AGBW02007806 OWR54532.1 CH477459 EAT40600.1 GAKP01008996 JAC49956.1 APCN01001644 GDHF01019860 JAI32454.1 JXUM01018539 JXUM01018540 JXUM01018541 JXUM01018542 KQ560515 KXJ82034.1 AXCM01008115 ATLV01011119 KE524642 KFB35854.1 GEBQ01006132 JAT33845.1 GECZ01003930 JAS65839.1 GBXI01016390 GBXI01008465 JAC97901.1 JAD05827.1 ADMH02000277 ETN67143.1 UFQT01000011 SSX17590.1 JRES01000498 KNC30773.1 GDHF01026519 JAI25795.1 AJVK01003621 KK852945 KDR13451.1 ODYU01009337 SOQ53691.1 GAMC01002610 JAC03946.1 CH963920 EDW77965.1 JXJN01018354 CH379061 EAL33235.2 CH479228 EDW35384.1 CH902620 EDV30671.1 CCAG010020516 KB632308 ERL92021.1 CH940654 EDW57625.1 AB056651 BAB33388.1 KK107348 EZA52208.1 OUUW01000006 SPP82440.1 JXUM01034205 JXUM01034206 KQ561016 KXJ80024.1 CM000361 EDX05165.1 QOIP01000010 RLU17935.1 CH916368 EDW03655.1 CH954177 EDV58058.1 CH933807 EDW11840.1 CM000157 EDW89713.1 CH954297 EDV47705.1 KOB75887.1 PZC72472.1 AE014134 AY058398 AAF53488.1 AAL13627.1 AGB93011.1 CM002910 KMY90417.1 CH480820 EDW54532.1 LBMM01006278 KMQ90745.1 CP012523 ALC39886.1 KQ434820 KZC07027.1 PCG80510.1 GFTR01005315 JAW11111.1 KQ971357 EFA08354.1 ACPB03003574 OWR54533.1 ALC39895.1 NIVC01002675 NIVC01001955 PAA56021.1 PAA62277.1 KQ982578 KYQ54626.1 NIVC01002189 PAA60139.1 KK854992 PTY20160.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000053268
UP000037510
UP000218220
+ More
UP000095301 UP000235965 UP000183832 UP000002320 UP000092461 UP000007062 UP000075901 UP000008820 UP000075920 UP000076407 UP000075880 UP000075840 UP000075903 UP000069272 UP000069940 UP000249989 UP000075883 UP000075885 UP000030765 UP000000673 UP000037069 UP000075881 UP000075884 UP000075902 UP000075900 UP000092462 UP000027135 UP000078200 UP000091820 UP000092443 UP000007798 UP000092460 UP000001819 UP000008744 UP000007801 UP000092444 UP000030742 UP000008792 UP000092445 UP000053097 UP000268350 UP000000304 UP000192221 UP000279307 UP000001070 UP000008711 UP000009192 UP000002282 UP000000803 UP000001292 UP000036403 UP000092553 UP000076502 UP000007266 UP000015103 UP000076408 UP000076420 UP000095280 UP000215902 UP000075809 UP000007875
UP000095301 UP000235965 UP000183832 UP000002320 UP000092461 UP000007062 UP000075901 UP000008820 UP000075920 UP000076407 UP000075880 UP000075840 UP000075903 UP000069272 UP000069940 UP000249989 UP000075883 UP000075885 UP000030765 UP000000673 UP000037069 UP000075881 UP000075884 UP000075902 UP000075900 UP000092462 UP000027135 UP000078200 UP000091820 UP000092443 UP000007798 UP000092460 UP000001819 UP000008744 UP000007801 UP000092444 UP000030742 UP000008792 UP000092445 UP000053097 UP000268350 UP000000304 UP000192221 UP000279307 UP000001070 UP000008711 UP000009192 UP000002282 UP000000803 UP000001292 UP000036403 UP000092553 UP000076502 UP000007266 UP000015103 UP000076408 UP000076420 UP000095280 UP000215902 UP000075809 UP000007875
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JW86
A0A0N1IBU9
A0A212F8N0
A0A194PJK5
A0A0L7LCK7
H9JWM1
+ More
A0A2A4K8G5 A0A1I8MQ14 A0A2W1BDL7 A0A2J7Q0W3 A0A194Q0S4 A0A1J1IHY3 A0A2A4KA34 A0A0L7LJZ3 A0A0N1IQ59 A0A1B6BY47 B0WGB8 A0A2W1BI80 A0A1B0CEK9 Q7Q108 A0A023ET07 A0A212FLC4 A0A182SIM5 Q171C6 A0A182W8M4 A0A182XHD2 A0A182JDH7 A0A034W300 A0A1I8JTJ3 A0A0K8V0M0 A0A182VFR4 A0A182F336 A0A182GU33 A0A182M6S8 A0A182PDW9 A0A084VD10 A0A1B6MD53 A0A1B6GTT4 A0A0A1X419 W5JV24 A0A336LID9 A0A0L0CED8 A0A182K1J2 A0A182NNY0 A0A182TIT4 A0A0K8UHJ6 A0A182R1T9 A0A1B0D6I8 A0A067QTU0 A0A1A9V1K0 A0A2H1WKT9 A0A182PEF9 A0A1A9WUB4 W8BRM4 A0A1A9Y8T0 B4N030 A0A1B0BQ23 Q29KE0 B4H9F2 B3ML13 A0A1B0FL25 U4UPD2 B4M8U9 Q9BLD7 A0A1A9ZWA8 A0A026W8B8 H9JWM2 A0A3B0K3C4 A0A182HDJ0 B4Q6I3 A0A1W4U4P9 A0A3L8DBQ9 B4JED5 B3N620 B4KE95 B4NY77 B3PA87 A0A0L7LKB4 A0A2W1BAS8 Q9V3M9 A0A0J9R2F1 B4I1X5 A0A0J7NDZ5 A0A0M5IZ51 A0A154P562 A0A2A4K9W1 A0A224XT22 D6WUN0 T1I5P5 A0A182Y659 A0A212FLB2 A0A0M4ES94 A0A2C9JZH2 A0A1I8FZ59 A0A267EN10 A0A151X2K1 A0A267EH03 H2YEY8 A0A2R7WMD4
A0A2A4K8G5 A0A1I8MQ14 A0A2W1BDL7 A0A2J7Q0W3 A0A194Q0S4 A0A1J1IHY3 A0A2A4KA34 A0A0L7LJZ3 A0A0N1IQ59 A0A1B6BY47 B0WGB8 A0A2W1BI80 A0A1B0CEK9 Q7Q108 A0A023ET07 A0A212FLC4 A0A182SIM5 Q171C6 A0A182W8M4 A0A182XHD2 A0A182JDH7 A0A034W300 A0A1I8JTJ3 A0A0K8V0M0 A0A182VFR4 A0A182F336 A0A182GU33 A0A182M6S8 A0A182PDW9 A0A084VD10 A0A1B6MD53 A0A1B6GTT4 A0A0A1X419 W5JV24 A0A336LID9 A0A0L0CED8 A0A182K1J2 A0A182NNY0 A0A182TIT4 A0A0K8UHJ6 A0A182R1T9 A0A1B0D6I8 A0A067QTU0 A0A1A9V1K0 A0A2H1WKT9 A0A182PEF9 A0A1A9WUB4 W8BRM4 A0A1A9Y8T0 B4N030 A0A1B0BQ23 Q29KE0 B4H9F2 B3ML13 A0A1B0FL25 U4UPD2 B4M8U9 Q9BLD7 A0A1A9ZWA8 A0A026W8B8 H9JWM2 A0A3B0K3C4 A0A182HDJ0 B4Q6I3 A0A1W4U4P9 A0A3L8DBQ9 B4JED5 B3N620 B4KE95 B4NY77 B3PA87 A0A0L7LKB4 A0A2W1BAS8 Q9V3M9 A0A0J9R2F1 B4I1X5 A0A0J7NDZ5 A0A0M5IZ51 A0A154P562 A0A2A4K9W1 A0A224XT22 D6WUN0 T1I5P5 A0A182Y659 A0A212FLB2 A0A0M4ES94 A0A2C9JZH2 A0A1I8FZ59 A0A267EN10 A0A151X2K1 A0A267EH03 H2YEY8 A0A2R7WMD4
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Nucleus
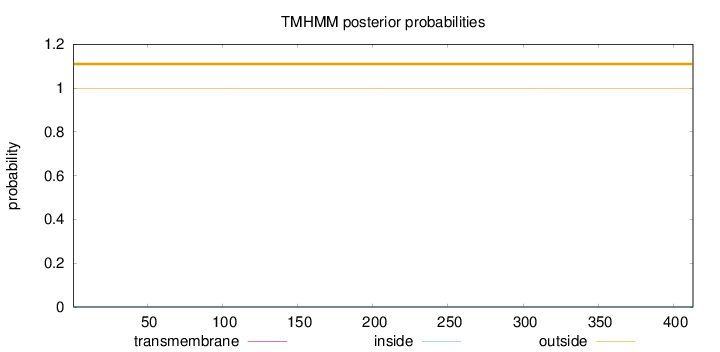
Length:
413
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00265
outside
1 - 413
Population Genetic Test Statistics
Pi
306.686287
Theta
192.415284
Tajima's D
1.869036
CLR
0
CSRT
0.862556872156392
Interpretation
Uncertain
