Gene
KWMTBOMO16371
Pre Gene Modal
BGIBMGA013799
Annotation
PREDICTED:_structural_maintenance_of_chromosomes_protein_5_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.548
Sequence
CDS
ATGTCGGAAACTTCTAATAATATAGGAGCGAGGCCTGGCTCCATATTCAGAATTGCGTTAGAAAACTTCCTTACATATAAAGAAGTAGAGCTGTTCCCAAACACATCACTGAACCTGATCATTGGTCCCAATGGAACAGGGAAGTCTACGTTTGTCTGTGCAATCATACTTGGACTATGCGGTAAGCCCCAACTTACGGGCCGATCAAAAAAGATAAATGACTATGTGCGCACTGGATGCGAAAAGGCAAAAATTGAGATAGAATTATATAAGGAACCGGGTGAAAGAAATGTGATTATAACGAGGACTTTCAATTTGCGAGGAGAGTCTTTGTGGGCGATTGACTATAGAAGTGTGAAAGAAAAAACTGTTCAGGATTTAGTAACTAGTTTGAAAATACAAGTGGACAACCTATGCCAGATGTTACCTCAAAACCGAGTCCAAGATTTCTGCCAAATGAACCCTCAGGAATTGCTCAAGAGAACGCTAGCAACGATCGGTGACCAGGAGGCCATAGGGCAAATGAACAATTTGGTAAATTACAGAACCGAACAAAGACAGTTGGACAACAAAATGCAAACTAACACCAAATTAATCGAGGAACAGATGACATTACATGAAACCTTAAAAAAGAAAATAGAAGCTATGCGACATCGAAACGAGTATGAAAAGAAAATTAGTATATGTGAAAAGAAGAAACTGTGGCTTCAATATAAACTGTTAAGTGCCAGAGCGGCGCAGACCGAGACTCGCATGGCAGAGGCTCTGAAAATAGTCAAGGCTAGTCACAAGAAGAAGCAGCCATTGGAAATGGCTGTACAGAGATTGAGAGAAGAAAAAATCAAATTGGAACAGGAAAGAGCAACAGGGAGTAGAAAAATAAATGAGCTAGCGGATAAAGTGAAAGAGCTGCTAAACGGCATCCAGGACTACGATAGACAGATCAAGGAGGTGGACCGCGCTTTTGAGGAGAAGCTGCAGAGGCACAGGAATAGAGAGAGAGAGTTGGTCGAGGCCAGAACGAAACTCGACAAACTGAAGAACGACAGAACTAGCCTCATTGAGAAATTTGGCGACGAGAAAATAGCAAGTGAGAAAGCAAGAGCGCTGAGGGAACGTATAAGCGAGATCCAATCACAAATCCACAGCATTCAGAGGAGCAAAGCCAGGAAGGAGATGGAATATGAACAGCAGACTCTACCCAACCTTAGGATATTGAAAAACAAATTAAGGAAATTAGAAGACGTCGAGACGAAGCGTTTAGAGACTTTGAAAGAATACAGCGAGGACACCTACAAGGCGCTGGAATGGCTCAACGAGAACAAGGAGTTGTTCCATGACCAGGTCTACGGGCCCATGATGCTCGAGCTGAAATTCACGCATCCGGAGACCGCCCGCTACCTCGAGTCGACGGTCTCCAGCAAGGACCTGGTCGCCTTCACGTTCCAGTCCAGCCGCGACATGAACCTCTTCCTGCAGAGGGTCCGCGAGGAGCTGCAGCTGCGCGCCGTCAGCGCCCTCTGCAGCCGCGCCGGGGACTTCAGCGCGCCCCACCACGACATCAGGGAACTCAGCTACCTCGGGTTCCAGAGGACCTTGGTGGAGGGCATAGCGGCGCCGGGCCCGGTGCTGCGGTACCTGTGCGAGCAGTACCAAGTCCACAGGATCCCCATCGGGAACGACCACACGTACCACCACAGCGAGAACGTGCCGGATCACGTCACGCTGTTTTTTACAACGCGGCACCGGTTCAGCGTGCGCGTGTCGCAGTACAGCGGCGCGCGCGCCAGCACGGTGGGCGAGGTGCGGCCGCCGCGCCTGCTTGCCAACGCGCGCGACGCCGACCTCATCCGCACCTGCGCGCTACAATTGAAAACATTAAATGCGACCTCGGAAAGCCAATTGGCCGCCATCAAGAGTCTGGGAGATCACATCTCGGAGCTGGAACAAAGGCAGGGCCAACTAAATAATGAATTGAAAGTTATTAAAGACAATGCATCAAAGATCGCAGAGATATCGTGCCAAATAAGAGTTCAGGCGAGGAAAGTGGAGGGCATCGAGAACGAGGAGTCCTTCGACGTCGCCGCCGAGTCGAGCCGCGCCGCCGCACTGCGGGCCCGCGCCGCCGCCGCCCAGCGCCGCCTGCACGCGGGGGGCGCGGGGGCGCTGCGCCGCCTGCACGCCGCCGCCGCCGCGCCCGCGCTGCTCAAGGTTAAACTGGAGATAATACGCGATGAGATCGTTCAACGGGAATCAGAATTGATCGATCTGAGAAACGAATTGGACGAAGCCAAGAGATCCTTGGAGCTGGTGCAGACGCAGCACGCTGAAGCTCAGTCGAGGGCGGCCGCCAAGCTGTCTCAGGTGCGGGCTCGCTGCGGCGCCGCGGACCCCGCCGACCCCGCCTCCCCCCTCGCGGCGCTGTTCCGCGACCTGCCGGACGAGCTGGACGCGCTGGCCGAGCGCTGCTACGAACTGCAGACCAGCGTCGACTGTATGGACAAGGAGACCGGGAACGTGATAAAGGATTACGAGGAATGCGAGAAAGCGATAGAGGCTCTCCAAGCTGAGAACAAGAATGTTGACCGACAGAACCAGACCTTGCTACGGCGGATGGACACGCTCAGCACGCAATGGCTGCCGACCGTCGAGAAACTTCTCCAGGAGATCGACAAACGTTTCGGCGAGATGTTCGCGTTGCTGAACTGCGCCGGCGAAGTGAGACTCGACAAAGGGAACAGCGACGAGGACTACGATAAGTACGGCGTGGTCGTACTGGTGAGGTTCCGGGAATCTGAGGAGTTGCAGCCGCTCTGCCGCAAGAGGCACTCCGGGGGGGAGCGCGCGCTCTCCATCGCGCTGTACCTCATGGCCCTGCAGAGGCTCTTCACAGTGCCCTTCAGATGCGTGGACGAAATAAATCAAGGCATGGACGAGATCAATGAACGCAAGATGTTCCAACTCCTGGTCCAAGTGACGAGTGAATGTGAGAACTCCCAGTACTTCCTGTTGACGCCCAAACTGCTGTCCAATCTGGAGTACAACGACAAGATAATGGTCCACACCATCATGAACGGGAAACACGTCATGAGATATAAAGAATGGGACCACGCCACGTTCGTGCAGAGGGCACGGCAGCTAAGCAGATCCTGA
Protein
MSETSNNIGARPGSIFRIALENFLTYKEVELFPNTSLNLIIGPNGTGKSTFVCAIILGLCGKPQLTGRSKKINDYVRTGCEKAKIEIELYKEPGERNVIITRTFNLRGESLWAIDYRSVKEKTVQDLVTSLKIQVDNLCQMLPQNRVQDFCQMNPQELLKRTLATIGDQEAIGQMNNLVNYRTEQRQLDNKMQTNTKLIEEQMTLHETLKKKIEAMRHRNEYEKKISICEKKKLWLQYKLLSARAAQTETRMAEALKIVKASHKKKQPLEMAVQRLREEKIKLEQERATGSRKINELADKVKELLNGIQDYDRQIKEVDRAFEEKLQRHRNRERELVEARTKLDKLKNDRTSLIEKFGDEKIASEKARALRERISEIQSQIHSIQRSKARKEMEYEQQTLPNLRILKNKLRKLEDVETKRLETLKEYSEDTYKALEWLNENKELFHDQVYGPMMLELKFTHPETARYLESTVSSKDLVAFTFQSSRDMNLFLQRVREELQLRAVSALCSRAGDFSAPHHDIRELSYLGFQRTLVEGIAAPGPVLRYLCEQYQVHRIPIGNDHTYHHSENVPDHVTLFFTTRHRFSVRVSQYSGARASTVGEVRPPRLLANARDADLIRTCALQLKTLNATSESQLAAIKSLGDHISELEQRQGQLNNELKVIKDNASKIAEISCQIRVQARKVEGIENEESFDVAAESSRAAALRARAAAAQRRLHAGGAGALRRLHAAAAAPALLKVKLEIIRDEIVQRESELIDLRNELDEAKRSLELVQTQHAEAQSRAAAKLSQVRARCGAADPADPASPLAALFRDLPDELDALAERCYELQTSVDCMDKETGNVIKDYEECEKAIEALQAENKNVDRQNQTLLRRMDTLSTQWLPTVEKLLQEIDKRFGEMFALLNCAGEVRLDKGNSDEDYDKYGVVVLVRFRESEELQPLCRKRHSGGERALSIALYLMALQRLFTVPFRCVDEINQGMDEINERKMFQLLVQVTSECENSQYFLLTPKLLSNLEYNDKIMVHTIMNGKHVMRYKEWDHATFVQRARQLSRS
Summary
Uniprot
A0A2H1W2E1
A0A212F8L8
A0A0N0P9C4
A0A067RH32
Q16RL3
A0A1Q3F742
+ More
A0A1Q3F712 A0A182HE17 A0A232FBU9 K7IQT1 A0A1S4FRP3 B0WYP3 A0A336KDQ1 A0A336MNE2 D6WRS7 E2A038 A0A3L8DIG5 A0A182VLR7 A0A182WX64 A0A1Y1N6V5 A0A182LNW4 A0A182HN42 A0A182RHR0 A0A182MXM2 Q7PV59 A0A2A3E9A1 A0A182TG61 W8BLT3 Q8I950 A0A034VL56 A0A084VEY7 A0A182QYD2 A0A1S3GY52 A0A182PUN3 W8C1Z6 A0A182FBF2 A0A182WEM4 A0A182YNH3 A0A182IWM0 A0A182KE75 A0A1V4KCV6 A0A182SRW3 W5JHJ0 F6Z1K2 A0A2C9K293 A0A1B0CS84 A0A3B4ZQ12 A0A3Q2U1Z1 H2ZZN0 A0A146MZB0 A0A3Q7WQI3 A0A3Q1EGV1 K7FR05 A0A146W8X2 A0A182LZF6 A0A1L8HP39 A0A2I0MFS0 A0A1B0GQF2 A0A091FLX5 A0A091WH05 U3IPC5 A0A3Q1G6T1 A0A3P8ULL6 A0A3P9BDT1 A0A0K8VQ12 A0A0K8W7R6
A0A1Q3F712 A0A182HE17 A0A232FBU9 K7IQT1 A0A1S4FRP3 B0WYP3 A0A336KDQ1 A0A336MNE2 D6WRS7 E2A038 A0A3L8DIG5 A0A182VLR7 A0A182WX64 A0A1Y1N6V5 A0A182LNW4 A0A182HN42 A0A182RHR0 A0A182MXM2 Q7PV59 A0A2A3E9A1 A0A182TG61 W8BLT3 Q8I950 A0A034VL56 A0A084VEY7 A0A182QYD2 A0A1S3GY52 A0A182PUN3 W8C1Z6 A0A182FBF2 A0A182WEM4 A0A182YNH3 A0A182IWM0 A0A182KE75 A0A1V4KCV6 A0A182SRW3 W5JHJ0 F6Z1K2 A0A2C9K293 A0A1B0CS84 A0A3B4ZQ12 A0A3Q2U1Z1 H2ZZN0 A0A146MZB0 A0A3Q7WQI3 A0A3Q1EGV1 K7FR05 A0A146W8X2 A0A182LZF6 A0A1L8HP39 A0A2I0MFS0 A0A1B0GQF2 A0A091FLX5 A0A091WH05 U3IPC5 A0A3Q1G6T1 A0A3P8ULL6 A0A3P9BDT1 A0A0K8VQ12 A0A0K8W7R6
Pubmed
EMBL
ODYU01005893
SOQ47249.1
AGBW02009696
OWR50096.1
KQ459547
KPJ00019.1
+ More
KK852639 KDR19631.1 CH477704 EAT37055.1 GFDL01011688 JAV23357.1 GFDL01011767 JAV23278.1 JXUM01130857 KQ567633 KXJ69342.1 NNAY01000475 OXU28125.1 DS232195 EDS37153.1 UFQS01000239 UFQT01000239 SSX01848.1 SSX22225.1 UFQT01001390 SSX30333.1 KQ971354 EFA05955.1 GL435551 EFN73171.1 QOIP01000007 RLU20141.1 GEZM01011169 JAV93613.1 APCN01001834 AAAB01008986 EAA00424.3 KZ288317 PBC28307.1 GAMC01006778 JAB99777.1 AJ535207 CAD59407.1 GAKP01016397 JAC42555.1 ATLV01012354 KE524784 KFB36531.1 AXCN02001064 GAMC01006779 JAB99776.1 LSYS01003700 OPJ82306.1 ADMH02001485 ETN62375.1 AAMC01047916 AAMC01047917 AAMC01047918 AAMC01047919 AAMC01047920 AJWK01025799 AFYH01125448 AFYH01125449 AFYH01125450 AFYH01125451 GCES01162312 JAQ24010.1 AGCU01194619 AGCU01194620 AGCU01194621 AGCU01194622 AGCU01194623 AGCU01194624 AGCU01194625 AGCU01194626 GCES01060801 JAR25522.1 AXCM01005319 CM004467 OCT97869.1 AKCR02000015 PKK28524.1 AJVK01007177 AJVK01007178 KL447131 KFO70159.1 KK735464 KFR14148.1 ADON01057353 ADON01057354 ADON01057355 ADON01057356 ADON01057357 ADON01057358 ADON01057359 ADON01057360 ADON01057361 ADON01057362 GDHF01011338 JAI40976.1 GDHF01005404 JAI46910.1
KK852639 KDR19631.1 CH477704 EAT37055.1 GFDL01011688 JAV23357.1 GFDL01011767 JAV23278.1 JXUM01130857 KQ567633 KXJ69342.1 NNAY01000475 OXU28125.1 DS232195 EDS37153.1 UFQS01000239 UFQT01000239 SSX01848.1 SSX22225.1 UFQT01001390 SSX30333.1 KQ971354 EFA05955.1 GL435551 EFN73171.1 QOIP01000007 RLU20141.1 GEZM01011169 JAV93613.1 APCN01001834 AAAB01008986 EAA00424.3 KZ288317 PBC28307.1 GAMC01006778 JAB99777.1 AJ535207 CAD59407.1 GAKP01016397 JAC42555.1 ATLV01012354 KE524784 KFB36531.1 AXCN02001064 GAMC01006779 JAB99776.1 LSYS01003700 OPJ82306.1 ADMH02001485 ETN62375.1 AAMC01047916 AAMC01047917 AAMC01047918 AAMC01047919 AAMC01047920 AJWK01025799 AFYH01125448 AFYH01125449 AFYH01125450 AFYH01125451 GCES01162312 JAQ24010.1 AGCU01194619 AGCU01194620 AGCU01194621 AGCU01194622 AGCU01194623 AGCU01194624 AGCU01194625 AGCU01194626 GCES01060801 JAR25522.1 AXCM01005319 CM004467 OCT97869.1 AKCR02000015 PKK28524.1 AJVK01007177 AJVK01007178 KL447131 KFO70159.1 KK735464 KFR14148.1 ADON01057353 ADON01057354 ADON01057355 ADON01057356 ADON01057357 ADON01057358 ADON01057359 ADON01057360 ADON01057361 ADON01057362 GDHF01011338 JAI40976.1 GDHF01005404 JAI46910.1
Proteomes
UP000007151
UP000053268
UP000027135
UP000008820
UP000069940
UP000249989
+ More
UP000215335 UP000002358 UP000002320 UP000007266 UP000000311 UP000279307 UP000075903 UP000076407 UP000075882 UP000075840 UP000075900 UP000075884 UP000007062 UP000242457 UP000075902 UP000030765 UP000075886 UP000085678 UP000075885 UP000069272 UP000075920 UP000076408 UP000075880 UP000075881 UP000190648 UP000075901 UP000000673 UP000008143 UP000076420 UP000092461 UP000261400 UP000265000 UP000008672 UP000286642 UP000257200 UP000007267 UP000075883 UP000186698 UP000053872 UP000092462 UP000053760 UP000053605 UP000016666 UP000265120 UP000265160
UP000215335 UP000002358 UP000002320 UP000007266 UP000000311 UP000279307 UP000075903 UP000076407 UP000075882 UP000075840 UP000075900 UP000075884 UP000007062 UP000242457 UP000075902 UP000030765 UP000075886 UP000085678 UP000075885 UP000069272 UP000075920 UP000076408 UP000075880 UP000075881 UP000190648 UP000075901 UP000000673 UP000008143 UP000076420 UP000092461 UP000261400 UP000265000 UP000008672 UP000286642 UP000257200 UP000007267 UP000075883 UP000186698 UP000053872 UP000092462 UP000053760 UP000053605 UP000016666 UP000265120 UP000265160
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A2H1W2E1
A0A212F8L8
A0A0N0P9C4
A0A067RH32
Q16RL3
A0A1Q3F742
+ More
A0A1Q3F712 A0A182HE17 A0A232FBU9 K7IQT1 A0A1S4FRP3 B0WYP3 A0A336KDQ1 A0A336MNE2 D6WRS7 E2A038 A0A3L8DIG5 A0A182VLR7 A0A182WX64 A0A1Y1N6V5 A0A182LNW4 A0A182HN42 A0A182RHR0 A0A182MXM2 Q7PV59 A0A2A3E9A1 A0A182TG61 W8BLT3 Q8I950 A0A034VL56 A0A084VEY7 A0A182QYD2 A0A1S3GY52 A0A182PUN3 W8C1Z6 A0A182FBF2 A0A182WEM4 A0A182YNH3 A0A182IWM0 A0A182KE75 A0A1V4KCV6 A0A182SRW3 W5JHJ0 F6Z1K2 A0A2C9K293 A0A1B0CS84 A0A3B4ZQ12 A0A3Q2U1Z1 H2ZZN0 A0A146MZB0 A0A3Q7WQI3 A0A3Q1EGV1 K7FR05 A0A146W8X2 A0A182LZF6 A0A1L8HP39 A0A2I0MFS0 A0A1B0GQF2 A0A091FLX5 A0A091WH05 U3IPC5 A0A3Q1G6T1 A0A3P8ULL6 A0A3P9BDT1 A0A0K8VQ12 A0A0K8W7R6
A0A1Q3F712 A0A182HE17 A0A232FBU9 K7IQT1 A0A1S4FRP3 B0WYP3 A0A336KDQ1 A0A336MNE2 D6WRS7 E2A038 A0A3L8DIG5 A0A182VLR7 A0A182WX64 A0A1Y1N6V5 A0A182LNW4 A0A182HN42 A0A182RHR0 A0A182MXM2 Q7PV59 A0A2A3E9A1 A0A182TG61 W8BLT3 Q8I950 A0A034VL56 A0A084VEY7 A0A182QYD2 A0A1S3GY52 A0A182PUN3 W8C1Z6 A0A182FBF2 A0A182WEM4 A0A182YNH3 A0A182IWM0 A0A182KE75 A0A1V4KCV6 A0A182SRW3 W5JHJ0 F6Z1K2 A0A2C9K293 A0A1B0CS84 A0A3B4ZQ12 A0A3Q2U1Z1 H2ZZN0 A0A146MZB0 A0A3Q7WQI3 A0A3Q1EGV1 K7FR05 A0A146W8X2 A0A182LZF6 A0A1L8HP39 A0A2I0MFS0 A0A1B0GQF2 A0A091FLX5 A0A091WH05 U3IPC5 A0A3Q1G6T1 A0A3P8ULL6 A0A3P9BDT1 A0A0K8VQ12 A0A0K8W7R6
PDB
5XEI
E-value=3.2146e-08,
Score=143
Ontologies
PANTHER
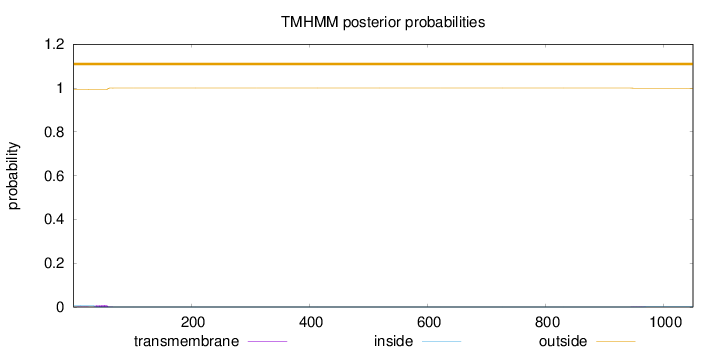
Topology
Length:
1050
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.18411
Exp number, first 60 AAs:
0.16256
Total prob of N-in:
0.00734
outside
1 - 1050
Population Genetic Test Statistics
Pi
308.695225
Theta
168.916278
Tajima's D
3.409309
CLR
0.199657
CSRT
0.991600419979001
Interpretation
Uncertain
