Gene
KWMTBOMO16370 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013746
Annotation
prophenoloxidase_activating_enzyme_precursor_[Bombyx_mori]
Full name
Phenoloxidase-activating enzyme
Alternative Name
Prophenoloxidase-activating enzyme
Location in the cell
Extracellular Reliability : 4.629
Sequence
CDS
ATGTTTTTAATTTGGACATTCATCGTGGCTGTTCTGGCGATCCAGACCAAAAGTGTTGTTGCACAATCATGCCGGACACCGAACGGACTTAACGGTAATTGCGTATCCGTGTATGAATGCCAGGCACTCCTGGCCATATTAAATAATCAGAGGAGAACCCAGCAAGATGAGAAGTTCTTGAGGGACTCCCAATGCGGCACCAAGAATTCAGTTCCTGCTGTTTGCTGTCCATGCAACGCAGCTGATGGTCAGCAAGGGAATTGTGTCAACATAAATTCATGTCCGTACGTTCTTCAATTGCTGAAGAATCCGAATGAAGCGAATCTAAATTACGTCCGGGGATCTGTTTGTCAAGGATCAGAACAACAGAGCATATGCTGTGTTACTGCGCCACAGTCGACGGCAGTGACCACGACCCCCAGACCGAAGCGGGTGCACGCCTGCCAGAGCGAGATGACTGCCACCCCCCCTAATCCGGAAGGGAAGTGTTGCGGCCGGGACATAGCCGTCGGAGACAAAATCGTTGGTGGTGCGCCCGCATCCATCGACTCGTATCCTTGGCTGGTGGTCATCGAGTATGTGAGGCTCGAGAGGACGATGCTGCTCTGTGGCGGGGCGCTCATCAGCGGGAAGTACGTGCTCACTGCCGGTCATTGCGTGAAAGGAGCGATCTTGGATGTAGGCACGCCGAAAACAGTGCGTCTCGGCGAGTACAACACGACGAACCCGGGCCGTGATTGCGTGTCCGTATCAGCGGGCGGCACGGATTGCACGGACCCATTGGTCAAGATAGGCATTGAGAAGACCATACCTCACCCCGACTATCAGCCCTATCATTTTCTTAGGAAACACGACATCGGGTTGATCAGGCTGCAAAGCATCGCGCCTTTTACAGATTTCATCAGGCCTATATGCTTGCCGAGTACAGATTACACAGTGAACCCGCCAAGTAAATTCGCTCTGACCGTGGCTGGCTGGGGACGATACTTGCAGTTCGACAATGGGACAGTTAGAAGCAGCAAGATCAAGCTTCACGTAACACTGCCTTTTGTACAACGAGACGTCTGCGAAGCAAACCAGAAACCATTGCGCAATGGCCAAAGAATAACATTGTGGAAGGGACAAATGTGTGCCGGCGGTGAAGCAGGTAAAGATTCTTGTAAGGGTGACTCGGGTGGTCCTCTTATGTATGAACATTCCAAAAAATACGAAGCAGTCGGTATTGTGAGCTTCGGACCCGAAAAGTGTGGCCAGATCGATATACCTGGAGTTTACACAAATGTTTATGAATACCTGCCATGGATACAAAATACAATTGAACCTTGA
Protein
MFLIWTFIVAVLAIQTKSVVAQSCRTPNGLNGNCVSVYECQALLAILNNQRRTQQDEKFLRDSQCGTKNSVPAVCCPCNAADGQQGNCVNINSCPYVLQLLKNPNEANLNYVRGSVCQGSEQQSICCVTAPQSTAVTTTPRPKRVHACQSEMTATPPNPEGKCCGRDIAVGDKIVGGAPASIDSYPWLVVIEYVRLERTMLLCGGALISGKYVLTAGHCVKGAILDVGTPKTVRLGEYNTTNPGRDCVSVSAGGTDCTDPLVKIGIEKTIPHPDYQPYHFLRKHDIGLIRLQSIAPFTDFIRPICLPSTDYTVNPPSKFALTVAGWGRYLQFDNGTVRSSKIKLHVTLPFVQRDVCEANQKPLRNGQRITLWKGQMCAGGEAGKDSCKGDSGGPLMYEHSKKYEAVGIVSFGPEKCGQIDIPGVYTNVYEYLPWIQNTIEP
Summary
Description
Endopeptidase with selective post-Arg cleavage site (PubMed:4197814, PubMed:4197815, PubMed:10066809, PubMed:7981662). Activates prophenoloxidase (PubMed:4197814, PubMed:4197815). Has a probable role in the melanization process as part of the innate immune response (Probable).
Biophysicochemical Properties
2.2 uM for prophenoloxidase (at pH 7.6 and 0 degrees Celsius)
1.43 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the absence of KCl)
0.77 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the presence of 0.025 M KCl)
1.2 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the presence of 0.004 M KCl)
0.35 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the presence of 0.5 M KCl)
0.145 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the presence of 0.5 M KCl)
0.709 mM for N-p-tosyl-L-arginine methyl ester hydrochloride (TAME) (in the presence of 0.5 M KCl)
1.43 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the absence of KCl)
0.77 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the presence of 0.025 M KCl)
1.2 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the presence of 0.004 M KCl)
0.35 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the presence of 0.5 M KCl)
0.145 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the presence of 0.5 M KCl)
0.709 mM for N-p-tosyl-L-arginine methyl ester hydrochloride (TAME) (in the presence of 0.5 M KCl)
Subunit
In the active form, heterodimer of a light chain and a heavy chain; disulfide-linked.
Similarity
Belongs to the peptidase S1 family. CLIP subfamily.
Belongs to the peptidase S1 family.
Belongs to the peptidase S1 family.
Keywords
Calcium
Complete proteome
Direct protein sequencing
Disulfide bond
Glycoprotein
Hydrolase
Metal-binding
Protease
Pyrrolidone carboxylic acid
Reference proteome
Serine protease
Signal
Zymogen
Feature
chain Phenoloxidase-activating enzyme light chain
Uniprot
Q9XXV0
Q2FAY5
Q8I917
A0A2A4JWX6
A0A290U627
Q49QW0
+ More
A1IIA5 Q6Y2X4 Q5DI98 Q5MPC0 Q2FAY7 Q5MPC2 Q2FAY6 I4DJE3 A0A126U9N5 A0A140EQ25 A0A0N1INY1 S4PHS5 Q2FAY4 A0A0N1IMQ8 A0A212EUU4 I4DS78 A0A0U1WT40 A0A173GPF0 A0A212F8R3 A0A0L7L0F2 H9JW83 V5GX21 D2XPX4 K7J980 A0A2A3EA55 A0A1Y1L107 A0A154P5I7 A0A212F8P6 A0A173GPF7 A0A195FEH3 U4U606 A0A0J7NWE6 A0A1I8N0W4 J3JYT9 A0A088A359 A0A158NI84 E2C8L2 A0A151WR74 A0A0C9QZC0 A0A1I8PP27 A0A195EG37 F4WPR9 A0A026WIN7 B1B5K1 A0JCK6 A0A1A9VW86 A0A3L8D7J0 A0A1W4XI92 A0A0N1PH79 B4N8B1 B3NYQ7 A0A034WRL1 N6TWW7 A0A0K8WKM9 W8CBZ2 A0A0K8U524 A0A0A1WGL4 A0A034VLF9 A0A0L7QXF0 A0A195C079 A0A0A1XFE0 A0A0M9A7E2 E2AV85 A0A1Y1KZL1 A0A3S2M731 A0A0L0CE89 Q17B76 A0A1A9X740 A0A1B0FQW4 A0A1I8JTB4 T1DTQ9 A0A0K8VB28 A0A182XUW4 A0A1Y1K9U3 A0A182VAF6 Q9Y1K6 A0A034V075 Q7QKL2 A0A0A1X7H3 A0A1Y1L1S6 A0A195AWL1 A0A1W4VMD7 W8BLL3 B3F5B1 A0A0A1XAL3 A0A1A9Z167 A0A182IIR9 A0A023EQV6 A0A182G580 A0A1Y1K3W7 A0A336LMT0 Q7QB73
A1IIA5 Q6Y2X4 Q5DI98 Q5MPC0 Q2FAY7 Q5MPC2 Q2FAY6 I4DJE3 A0A126U9N5 A0A140EQ25 A0A0N1INY1 S4PHS5 Q2FAY4 A0A0N1IMQ8 A0A212EUU4 I4DS78 A0A0U1WT40 A0A173GPF0 A0A212F8R3 A0A0L7L0F2 H9JW83 V5GX21 D2XPX4 K7J980 A0A2A3EA55 A0A1Y1L107 A0A154P5I7 A0A212F8P6 A0A173GPF7 A0A195FEH3 U4U606 A0A0J7NWE6 A0A1I8N0W4 J3JYT9 A0A088A359 A0A158NI84 E2C8L2 A0A151WR74 A0A0C9QZC0 A0A1I8PP27 A0A195EG37 F4WPR9 A0A026WIN7 B1B5K1 A0JCK6 A0A1A9VW86 A0A3L8D7J0 A0A1W4XI92 A0A0N1PH79 B4N8B1 B3NYQ7 A0A034WRL1 N6TWW7 A0A0K8WKM9 W8CBZ2 A0A0K8U524 A0A0A1WGL4 A0A034VLF9 A0A0L7QXF0 A0A195C079 A0A0A1XFE0 A0A0M9A7E2 E2AV85 A0A1Y1KZL1 A0A3S2M731 A0A0L0CE89 Q17B76 A0A1A9X740 A0A1B0FQW4 A0A1I8JTB4 T1DTQ9 A0A0K8VB28 A0A182XUW4 A0A1Y1K9U3 A0A182VAF6 Q9Y1K6 A0A034V075 Q7QKL2 A0A0A1X7H3 A0A1Y1L1S6 A0A195AWL1 A0A1W4VMD7 W8BLL3 B3F5B1 A0A0A1XAL3 A0A1A9Z167 A0A182IIR9 A0A023EQV6 A0A182G580 A0A1Y1K3W7 A0A336LMT0 Q7QB73
EC Number
3.4.21.-
Pubmed
10066809
19121390
4197814
4197815
7981662
16324822
+ More
12456683 28953952 17307003 14505699 16033436 15944088 22651552 26354079 23622113 22118469 26227816 20075255 28004739 23537049 25315136 22516182 21347285 20798317 21719571 24508170 18195005 30249741 17994087 25348373 24495485 25830018 26108605 17510324 10646969 12364791 14747013 17210077 24945155 26483478
12456683 28953952 17307003 14505699 16033436 15944088 22651552 26354079 23622113 22118469 26227816 20075255 28004739 23537049 25315136 22516182 21347285 20798317 21719571 24508170 18195005 30249741 17994087 25348373 24495485 25830018 26108605 17510324 10646969 12364791 14747013 17210077 24945155 26483478
EMBL
AB009670
BABH01025554
BAA76308.1
DQ115323
AAZ91696.1
AY077643
+ More
AAL76085.1 NWSH01000467 PCG76208.1 KY680241 ATD13322.1 AY677082 AAW24481.1 AB264091 BAF43530.1 AY188445 AAO74570.1 AH014670 AY789466 AAX18637.1 AY672790 AAV91012.1 AAZ91694.1 AY672788 AAV91010.1 AAZ91695.1 AK401411 BAM18033.1 KT751522 AML39475.1 KT751521 AML39474.1 KQ460936 KPJ10700.1 GAIX01000374 JAA92186.1 DQ115324 AAZ91697.1 KQ459547 KPJ00023.1 AGBW02012316 OWR45231.1 AK405456 BAM20768.1 JQ581598 AFX82594.1 KT006138 ANH58163.1 AGBW02009696 OWR50099.1 JTDY01003981 KOB68749.1 BABH01025543 BABH01025544 GALX01002154 JAB66312.1 GU229936 ADA84072.3 AAZX01002213 KZ288322 PBC28172.1 GEZM01073843 JAV64787.1 KQ434809 KZC06390.1 OWR50098.1 KT006137 ANH58162.1 KQ981636 KYN38803.1 KB631684 ERL85365.1 LBMM01001207 KMQ96720.1 BT128420 AEE63377.1 ADTU01016095 GL453666 EFN75742.1 KQ982813 KYQ50307.1 GBYB01001065 JAG70832.1 KQ978957 KYN27230.1 GL888255 EGI63810.1 KK107185 EZA55838.1 AB363980 BAG14262.2 AB282644 BAF36824.1 QOIP01000012 RLU16063.1 KQ459999 KPJ18497.1 CH964232 EDW81362.1 CH954181 EDV48170.1 GAKP01002127 JAC56825.1 APGK01057809 KB741282 ENN70807.1 GDHF01000894 JAI51420.1 GAMC01000094 JAC06462.1 GDHF01030580 JAI21734.1 GBXI01016739 GBXI01006351 JAC97552.1 JAD07941.1 GAKP01015995 JAC42957.1 KQ414704 KOC63285.1 KQ978501 KYM93571.1 GBXI01004615 JAD09677.1 KQ435719 KOX78755.1 GL443059 EFN62639.1 GEZM01073844 JAV64786.1 RSAL01000017 RVE52975.1 JRES01000503 KNC30561.1 CH477325 EAT43491.1 CCAG010023843 APCN01002147 GAMD01000809 JAB00782.1 GDHF01016160 JAI36154.1 GEZM01092820 JAV56445.1 AF117749 AAD38335.1 GAKP01023416 JAC35542.1 AAAB01008794 EAA03566.2 GBXI01007200 JAD07092.1 GEZM01066955 JAV67623.1 KQ976725 KYM76561.1 GAMC01016016 JAB90539.1 EF614332 ABU98654.1 GBXI01006130 JAD08162.1 APCN01002556 GAPW01002093 JAC11505.1 JXUM01042955 JXUM01042956 KQ561344 KXJ78893.1 GEZM01097973 JAV54026.1 UFQT01000091 SSX19534.1 AAAB01008880 EAA08507.4
AAL76085.1 NWSH01000467 PCG76208.1 KY680241 ATD13322.1 AY677082 AAW24481.1 AB264091 BAF43530.1 AY188445 AAO74570.1 AH014670 AY789466 AAX18637.1 AY672790 AAV91012.1 AAZ91694.1 AY672788 AAV91010.1 AAZ91695.1 AK401411 BAM18033.1 KT751522 AML39475.1 KT751521 AML39474.1 KQ460936 KPJ10700.1 GAIX01000374 JAA92186.1 DQ115324 AAZ91697.1 KQ459547 KPJ00023.1 AGBW02012316 OWR45231.1 AK405456 BAM20768.1 JQ581598 AFX82594.1 KT006138 ANH58163.1 AGBW02009696 OWR50099.1 JTDY01003981 KOB68749.1 BABH01025543 BABH01025544 GALX01002154 JAB66312.1 GU229936 ADA84072.3 AAZX01002213 KZ288322 PBC28172.1 GEZM01073843 JAV64787.1 KQ434809 KZC06390.1 OWR50098.1 KT006137 ANH58162.1 KQ981636 KYN38803.1 KB631684 ERL85365.1 LBMM01001207 KMQ96720.1 BT128420 AEE63377.1 ADTU01016095 GL453666 EFN75742.1 KQ982813 KYQ50307.1 GBYB01001065 JAG70832.1 KQ978957 KYN27230.1 GL888255 EGI63810.1 KK107185 EZA55838.1 AB363980 BAG14262.2 AB282644 BAF36824.1 QOIP01000012 RLU16063.1 KQ459999 KPJ18497.1 CH964232 EDW81362.1 CH954181 EDV48170.1 GAKP01002127 JAC56825.1 APGK01057809 KB741282 ENN70807.1 GDHF01000894 JAI51420.1 GAMC01000094 JAC06462.1 GDHF01030580 JAI21734.1 GBXI01016739 GBXI01006351 JAC97552.1 JAD07941.1 GAKP01015995 JAC42957.1 KQ414704 KOC63285.1 KQ978501 KYM93571.1 GBXI01004615 JAD09677.1 KQ435719 KOX78755.1 GL443059 EFN62639.1 GEZM01073844 JAV64786.1 RSAL01000017 RVE52975.1 JRES01000503 KNC30561.1 CH477325 EAT43491.1 CCAG010023843 APCN01002147 GAMD01000809 JAB00782.1 GDHF01016160 JAI36154.1 GEZM01092820 JAV56445.1 AF117749 AAD38335.1 GAKP01023416 JAC35542.1 AAAB01008794 EAA03566.2 GBXI01007200 JAD07092.1 GEZM01066955 JAV67623.1 KQ976725 KYM76561.1 GAMC01016016 JAB90539.1 EF614332 ABU98654.1 GBXI01006130 JAD08162.1 APCN01002556 GAPW01002093 JAC11505.1 JXUM01042955 JXUM01042956 KQ561344 KXJ78893.1 GEZM01097973 JAV54026.1 UFQT01000091 SSX19534.1 AAAB01008880 EAA08507.4
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000002358 UP000242457 UP000076502 UP000078541 UP000030742 UP000036403 UP000095301 UP000005203 UP000005205 UP000008237 UP000075809 UP000095300 UP000078492 UP000007755 UP000053097 UP000078200 UP000279307 UP000192223 UP000007798 UP000008711 UP000019118 UP000053825 UP000078542 UP000053105 UP000000311 UP000283053 UP000037069 UP000008820 UP000092443 UP000092444 UP000075840 UP000076407 UP000075903 UP000007062 UP000078540 UP000192221 UP000092445 UP000069940 UP000249989
UP000002358 UP000242457 UP000076502 UP000078541 UP000030742 UP000036403 UP000095301 UP000005203 UP000005205 UP000008237 UP000075809 UP000095300 UP000078492 UP000007755 UP000053097 UP000078200 UP000279307 UP000192223 UP000007798 UP000008711 UP000019118 UP000053825 UP000078542 UP000053105 UP000000311 UP000283053 UP000037069 UP000008820 UP000092443 UP000092444 UP000075840 UP000076407 UP000075903 UP000007062 UP000078540 UP000192221 UP000092445 UP000069940 UP000249989
Interpro
Gene 3D
CDD
ProteinModelPortal
Q9XXV0
Q2FAY5
Q8I917
A0A2A4JWX6
A0A290U627
Q49QW0
+ More
A1IIA5 Q6Y2X4 Q5DI98 Q5MPC0 Q2FAY7 Q5MPC2 Q2FAY6 I4DJE3 A0A126U9N5 A0A140EQ25 A0A0N1INY1 S4PHS5 Q2FAY4 A0A0N1IMQ8 A0A212EUU4 I4DS78 A0A0U1WT40 A0A173GPF0 A0A212F8R3 A0A0L7L0F2 H9JW83 V5GX21 D2XPX4 K7J980 A0A2A3EA55 A0A1Y1L107 A0A154P5I7 A0A212F8P6 A0A173GPF7 A0A195FEH3 U4U606 A0A0J7NWE6 A0A1I8N0W4 J3JYT9 A0A088A359 A0A158NI84 E2C8L2 A0A151WR74 A0A0C9QZC0 A0A1I8PP27 A0A195EG37 F4WPR9 A0A026WIN7 B1B5K1 A0JCK6 A0A1A9VW86 A0A3L8D7J0 A0A1W4XI92 A0A0N1PH79 B4N8B1 B3NYQ7 A0A034WRL1 N6TWW7 A0A0K8WKM9 W8CBZ2 A0A0K8U524 A0A0A1WGL4 A0A034VLF9 A0A0L7QXF0 A0A195C079 A0A0A1XFE0 A0A0M9A7E2 E2AV85 A0A1Y1KZL1 A0A3S2M731 A0A0L0CE89 Q17B76 A0A1A9X740 A0A1B0FQW4 A0A1I8JTB4 T1DTQ9 A0A0K8VB28 A0A182XUW4 A0A1Y1K9U3 A0A182VAF6 Q9Y1K6 A0A034V075 Q7QKL2 A0A0A1X7H3 A0A1Y1L1S6 A0A195AWL1 A0A1W4VMD7 W8BLL3 B3F5B1 A0A0A1XAL3 A0A1A9Z167 A0A182IIR9 A0A023EQV6 A0A182G580 A0A1Y1K3W7 A0A336LMT0 Q7QB73
A1IIA5 Q6Y2X4 Q5DI98 Q5MPC0 Q2FAY7 Q5MPC2 Q2FAY6 I4DJE3 A0A126U9N5 A0A140EQ25 A0A0N1INY1 S4PHS5 Q2FAY4 A0A0N1IMQ8 A0A212EUU4 I4DS78 A0A0U1WT40 A0A173GPF0 A0A212F8R3 A0A0L7L0F2 H9JW83 V5GX21 D2XPX4 K7J980 A0A2A3EA55 A0A1Y1L107 A0A154P5I7 A0A212F8P6 A0A173GPF7 A0A195FEH3 U4U606 A0A0J7NWE6 A0A1I8N0W4 J3JYT9 A0A088A359 A0A158NI84 E2C8L2 A0A151WR74 A0A0C9QZC0 A0A1I8PP27 A0A195EG37 F4WPR9 A0A026WIN7 B1B5K1 A0JCK6 A0A1A9VW86 A0A3L8D7J0 A0A1W4XI92 A0A0N1PH79 B4N8B1 B3NYQ7 A0A034WRL1 N6TWW7 A0A0K8WKM9 W8CBZ2 A0A0K8U524 A0A0A1WGL4 A0A034VLF9 A0A0L7QXF0 A0A195C079 A0A0A1XFE0 A0A0M9A7E2 E2AV85 A0A1Y1KZL1 A0A3S2M731 A0A0L0CE89 Q17B76 A0A1A9X740 A0A1B0FQW4 A0A1I8JTB4 T1DTQ9 A0A0K8VB28 A0A182XUW4 A0A1Y1K9U3 A0A182VAF6 Q9Y1K6 A0A034V075 Q7QKL2 A0A0A1X7H3 A0A1Y1L1S6 A0A195AWL1 A0A1W4VMD7 W8BLL3 B3F5B1 A0A0A1XAL3 A0A1A9Z167 A0A182IIR9 A0A023EQV6 A0A182G580 A0A1Y1K3W7 A0A336LMT0 Q7QB73
PDB
2XXL
E-value=5.88573e-43,
Score=439
Ontologies
KEGG
GO
Topology
SignalP
Position: 1 - 21,
Likelihood: 0.998347
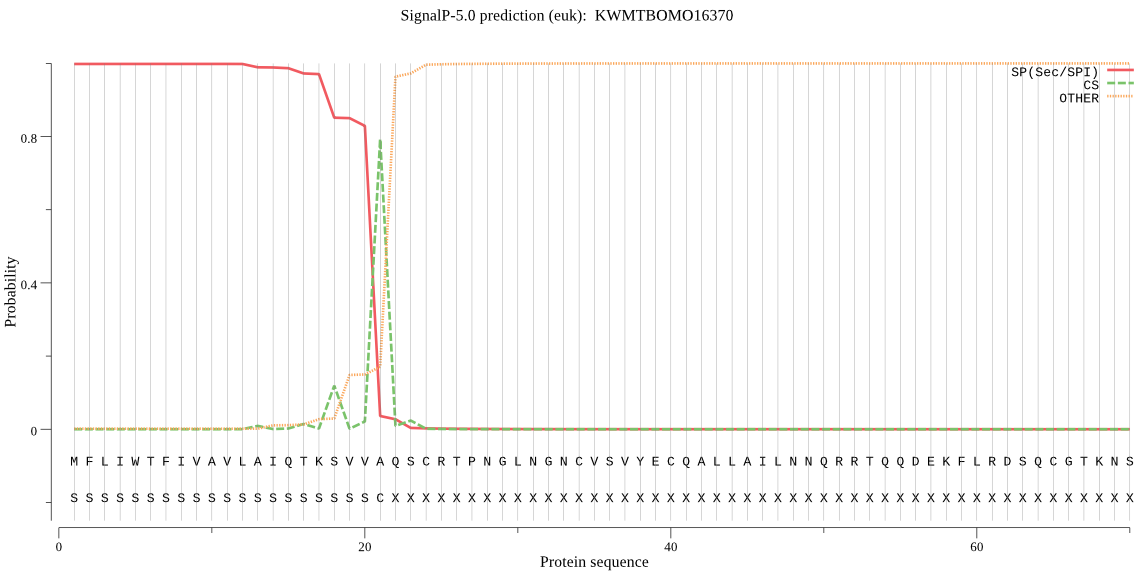
Length:
441
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.25092
Exp number, first 60 AAs:
0.24135
Total prob of N-in:
0.01256
outside
1 - 441

Population Genetic Test Statistics
Pi
215.33969
Theta
165.337877
Tajima's D
0.591947
CLR
0.802715
CSRT
0.542222888855557
Interpretation
Uncertain
