Gene
KWMTBOMO16366
Pre Gene Modal
BGIBMGA013797
Annotation
PREDICTED:_serine_protease_easter-like_[Bombyx_mori]
Full name
Phenoloxidase-activating enzyme
+ More
Melanization protease 1
Serine protease easter
Melanization protease 1
Serine protease easter
Alternative Name
Prophenoloxidase-activating enzyme
Location in the cell
Extracellular Reliability : 3.618
Sequence
CDS
ATGTCTCCGTTGTTGTTTACGACAATCGTAGCGGCTTATTTATGCGGCGCAAACGGACAGTCAGCGTGCACGGCCTCCGGTGGCGGTCAGGGAACGTGTGTGGATATCTACAAATGCGAGCCATTGCTGGCCCTCGTTAGAAAGCCTGATCGTACTCAACAGGATCTGGAAGCTCTCAAGAAATCTCAATGCGGGTTTCAGGGAGGGAAGCCATTGGTATGCTGTCCAAGTGAATGTCACACACCGGATGGTAAACCAGGAAAATGTATAAACATATACTCTTGCGAACACCTCACTCAAATGTTGAGGCCTCCGACCTCGAAGGAAAACATGTTATACGTACAAAACTCCAGGTGTAAAAGTGCAGATCAATACAGTGTATGCTGTGGCCCAGCGCCTTTAGCCACACCCCCTCCTCCAGCGGGCAGCTGCAGGGCGACTGTTTCGGCATTACTACCCAACCCAGCGGACTCTTGCTGTGGTCTAGACGCCAACGTTGGAAACAAAATCGTTGGTGGAACAGCGACTGCTATCGACCAATACCCGTGGCTGATAGTCATCGAGTATGTCAGTGATAACAGGATCAAATTATTGTGTGGTGGCTCGCTGATAAGCGGTAGATATGTTCTTACAGCTGGCCACTGCATAGCCGGAGCGGTCCTTCAAATCGGAAAACCTACCAACATTAGACTCGGCGAATACGACACTGCTAACAGCGGACCGGATTGTGTGGTTGTCGAAGCCGGAGGCACGGACTGCACGGAAGATATAGTAGTCATACCCATTGAACAGATCATACCCCATCCGGAATATAATCCTGTCAGCCCTCAAAGGAAGAACGATATCGCTTTGATCAGAATGAGTCAAATGGCTCCTTATACTGATTTTATCAGACCGATCTGCCTTCCATCGGCGGACATTACAGTGAACACACCAAATAAAACAGTATTCTACGCCGCCGGTTGGGGTGCAGTGAGCACGAACCAGATCAGCAGCGATGTCAAATCCCACGTAGACTTGCCTTACATTCCCCACAGTATTTGCCAACCAGCGTACTCGGTGCAAAACCGCAAAATAACTTTATGGAACAAACAGATTTGCGCTGGCGGTGTTCCGGGCAAAGACTCTTGCAAGGGCGACTCCGGTGGGCCATTGATGTACGAGAACGGTAGACAATATGAAGTCGTCGGTGTGGTCAGTTTTGGCCCAACGCCGTGCGGTCTTCCCGACATTCCGGGCGTTTACACCAAAGTCTACGAATACCTCGACTGGATCAGGAGCACGGTCAAATCATAG
Protein
MSPLLFTTIVAAYLCGANGQSACTASGGGQGTCVDIYKCEPLLALVRKPDRTQQDLEALKKSQCGFQGGKPLVCCPSECHTPDGKPGKCINIYSCEHLTQMLRPPTSKENMLYVQNSRCKSADQYSVCCGPAPLATPPPPAGSCRATVSALLPNPADSCCGLDANVGNKIVGGTATAIDQYPWLIVIEYVSDNRIKLLCGGSLISGRYVLTAGHCIAGAVLQIGKPTNIRLGEYDTANSGPDCVVVEAGGTDCTEDIVVIPIEQIIPHPEYNPVSPQRKNDIALIRMSQMAPYTDFIRPICLPSADITVNTPNKTVFYAAGWGAVSTNQISSDVKSHVDLPYIPHSICQPAYSVQNRKITLWNKQICAGGVPGKDSCKGDSGGPLMYENGRQYEVVGVVSFGPTPCGLPDIPGVYTKVYEYLDWIRSTVKS
Summary
Description
Endopeptidase with selective post-Arg cleavage site (PubMed:4197814, PubMed:4197815, PubMed:10066809, PubMed:7981662). Activates prophenoloxidase (PubMed:4197814, PubMed:4197815). Has a probable role in the melanization process as part of the innate immune response (Probable).
Serine protease which plays an essential role in the melanization immune response by acting downstream of sp7 to activate prophenoloxidase (PPO1) (PubMed:16861233). May function in diverse Hayan-dependent PPO1-activating cascades that are negatively controlled by different serpin proteins; Spn27A in the hemolymph and Spn77BA in the trachea (PubMed:18854145, PubMed:16861233). Regulation of melanization and PPO1 activation appears to be largely independent of the Toll signaling pathway (PubMed:16861233).
Component of the extracellular signaling pathway that establishes the dorsal-ventral pathway of the embryo (PubMed:9477324, PubMed:2107028, PubMed:12493753). Three proteases; ndl, gd and snk process easter to create active easter (PubMed:9477324). Active easter defines cell identities along the dorsal-ventral continuum by activating the spz ligand for the Tl receptor in the ventral region of the embryo (PubMed:9477324).
Serine protease which plays an essential role in the melanization immune response by acting downstream of sp7 to activate prophenoloxidase (PPO1) (PubMed:16861233). May function in diverse Hayan-dependent PPO1-activating cascades that are negatively controlled by different serpin proteins; Spn27A in the hemolymph and Spn77BA in the trachea (PubMed:18854145, PubMed:16861233). Regulation of melanization and PPO1 activation appears to be largely independent of the Toll signaling pathway (PubMed:16861233).
Component of the extracellular signaling pathway that establishes the dorsal-ventral pathway of the embryo (PubMed:9477324, PubMed:2107028, PubMed:12493753). Three proteases; ndl, gd and snk process easter to create active easter (PubMed:9477324). Active easter defines cell identities along the dorsal-ventral continuum by activating the spz ligand for the Tl receptor in the ventral region of the embryo (PubMed:9477324).
Biophysicochemical Properties
2.2 uM for prophenoloxidase (at pH 7.6 and 0 degrees Celsius)
1.43 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the absence of KCl)
0.77 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the presence of 0.025 M KCl)
1.2 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the presence of 0.004 M KCl)
0.35 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the presence of 0.5 M KCl)
0.145 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the presence of 0.5 M KCl)
0.709 mM for N-p-tosyl-L-arginine methyl ester hydrochloride (TAME) (in the presence of 0.5 M KCl)
1.43 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the absence of KCl)
0.77 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the presence of 0.025 M KCl)
1.2 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the presence of 0.004 M KCl)
0.35 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the presence of 0.5 M KCl)
0.145 mM for N-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) (in the presence of 0.5 M KCl)
0.709 mM for N-p-tosyl-L-arginine methyl ester hydrochloride (TAME) (in the presence of 0.5 M KCl)
Subunit
In the active form, heterodimer of a light chain and a heavy chain; disulfide-linked.
Similarity
Belongs to the peptidase S1 family.
Belongs to the peptidase S1 family. CLIP subfamily.
Belongs to the peptidase S1 family. CLIP subfamily.
Keywords
Calcium
Complete proteome
Direct protein sequencing
Disulfide bond
Glycoprotein
Hydrolase
Metal-binding
Protease
Pyrrolidone carboxylic acid
Reference proteome
Serine protease
Signal
Zymogen
Alternative splicing
Developmental protein
Secreted
Feature
chain Phenoloxidase-activating enzyme light chain
propeptide Activation peptide
splice variant In isoform A.
propeptide Activation peptide
splice variant In isoform A.
Uniprot
Q6Y2X4
Q5DI98
A0A2A4JWX6
A0A290U627
Q49QW0
A0A126U9N5
+ More
H9JW83 I4DJE3 A0A140EQ25 S4PHS5 A0A0N1INY1 A0A0N1IMQ8 A0A212EUU4 Q9XXV0 A0A0U1WT40 A0A212F8R3 A0A173GPF0 Q2FAY5 Q8I917 A1IIA5 I4DS78 Q5MPC0 Q2FAY6 Q2FAY4 Q2FAY7 Q5MPC2 A0A0L7L0F2 A0A126GUP6-3 A0A126GUP6-2 A0A173GPF7 A0A126GUP6 D2XPX4 W8CBZ2 B3P1Y6 A0A3B0K246 A0A3B0K4S8 A0A154P5I7 A0A1W4VLE8 A0A1J1J5N3 A0A1Y1L1S6 V5GX21 A0A2A3EA55 B4QUZ8 B0WQE7 J3JXM3 A0A034WRL1 A0A1I8PP27 A0A1I8NE97 A0A0A1WGL4 Q17B76 A0A182G580 A0A023EQV6 B3P0U7 B4PTQ4 A0A1Q3FHX1 A0A0M4EWB3 A0A0K8WKM9 Q295E4 A0A1I8N0W4 A0A0K8U524 A0A034VLF9 A0A182G5Z0 P13582 A0A034V0K7 K7J980 A0A336LMT0 A0A0K8VF53 B3MSM9 B4GM57 B4MBJ7 A0A1W4VIJ6 A0A0A1X7H3 A0A182SX76 B4QYJ5 B4KCX7 E2C8L2 A0A1B0B4Z6 D3TMC5 B4HDX9 A0A1B0FFU3 A0A3L8D7J0 A0A026WIN7 Q5MPC6 A0A1A9YQ00 B4PS51 A0A034V075 A0A0A1WU83 A0A1Y1LA41 A0A1A9UFJ1 A0A1Y1LA48 A0A1Y1K3W7 A0A3B0KH63 A0A0C9QZC0 B4N7Z2 A0A182IIR9 Q7QB73 A0A2Y9D4S9 A0A1Q3FC43
H9JW83 I4DJE3 A0A140EQ25 S4PHS5 A0A0N1INY1 A0A0N1IMQ8 A0A212EUU4 Q9XXV0 A0A0U1WT40 A0A212F8R3 A0A173GPF0 Q2FAY5 Q8I917 A1IIA5 I4DS78 Q5MPC0 Q2FAY6 Q2FAY4 Q2FAY7 Q5MPC2 A0A0L7L0F2 A0A126GUP6-3 A0A126GUP6-2 A0A173GPF7 A0A126GUP6 D2XPX4 W8CBZ2 B3P1Y6 A0A3B0K246 A0A3B0K4S8 A0A154P5I7 A0A1W4VLE8 A0A1J1J5N3 A0A1Y1L1S6 V5GX21 A0A2A3EA55 B4QUZ8 B0WQE7 J3JXM3 A0A034WRL1 A0A1I8PP27 A0A1I8NE97 A0A0A1WGL4 Q17B76 A0A182G580 A0A023EQV6 B3P0U7 B4PTQ4 A0A1Q3FHX1 A0A0M4EWB3 A0A0K8WKM9 Q295E4 A0A1I8N0W4 A0A0K8U524 A0A034VLF9 A0A182G5Z0 P13582 A0A034V0K7 K7J980 A0A336LMT0 A0A0K8VF53 B3MSM9 B4GM57 B4MBJ7 A0A1W4VIJ6 A0A0A1X7H3 A0A182SX76 B4QYJ5 B4KCX7 E2C8L2 A0A1B0B4Z6 D3TMC5 B4HDX9 A0A1B0FFU3 A0A3L8D7J0 A0A026WIN7 Q5MPC6 A0A1A9YQ00 B4PS51 A0A034V075 A0A0A1WU83 A0A1Y1LA41 A0A1A9UFJ1 A0A1Y1LA48 A0A1Y1K3W7 A0A3B0KH63 A0A0C9QZC0 B4N7Z2 A0A182IIR9 Q7QB73 A0A2Y9D4S9 A0A1Q3FC43
EC Number
3.4.21.-
Pubmed
14505699
16033436
28953952
19121390
22651552
23622113
+ More
26354079 22118469 10066809 4197814 4197815 7981662 16324822 12456683 17307003 15944088 26227816 10731132 12537572 10731138 16861233 18854145 22227521 24495485 17994087 28004739 22516182 25348373 25315136 25830018 17510324 26483478 24945155 17550304 15632085 2492450 9477324 9533958 2107028 12493753 20075255 20798317 20353571 30249741 24508170 12364791 14747013 17210077
26354079 22118469 10066809 4197814 4197815 7981662 16324822 12456683 17307003 15944088 26227816 10731132 12537572 10731138 16861233 18854145 22227521 24495485 17994087 28004739 22516182 25348373 25315136 25830018 17510324 26483478 24945155 17550304 15632085 2492450 9477324 9533958 2107028 12493753 20075255 20798317 20353571 30249741 24508170 12364791 14747013 17210077
EMBL
AY188445
AAO74570.1
AH014670
AY789466
AAX18637.1
NWSH01000467
+ More
PCG76208.1 KY680241 ATD13322.1 AY677082 AAW24481.1 KT751522 AML39475.1 BABH01025543 BABH01025544 AK401411 BAM18033.1 KT751521 AML39474.1 GAIX01000374 JAA92186.1 KQ460936 KPJ10700.1 KQ459547 KPJ00023.1 AGBW02012316 OWR45231.1 AB009670 BABH01025554 BAA76308.1 JQ581598 AFX82594.1 AGBW02009696 OWR50099.1 KT006138 ANH58163.1 DQ115323 AAZ91696.1 AY077643 AAL76085.1 AB264091 BAF43530.1 AK405456 BAM20768.1 AY672790 AAV91012.1 AAZ91695.1 DQ115324 AAZ91697.1 AAZ91694.1 AY672788 AAV91010.1 JTDY01003981 KOB68749.1 AE014297 AF145611 BT125977 AAD38586.1 ADX35956.1 KT006137 ANH58162.1 GU229936 ADA84072.3 GAMC01000094 JAC06462.1 CH954181 EDV47759.1 OUUW01000014 SPP88304.1 SPP88303.1 KQ434809 KZC06390.1 CVRI01000070 CRL07282.1 GEZM01066955 JAV67623.1 GALX01002154 JAB66312.1 KZ288322 PBC28172.1 CM000364 EDX11579.1 DS232039 EDS32818.1 BT127992 AEE62954.1 GAKP01002127 JAC56825.1 GBXI01016739 GBXI01006351 JAC97552.1 JAD07941.1 CH477325 EAT43491.1 JXUM01042955 JXUM01042956 KQ561344 KXJ78893.1 GAPW01002093 JAC11505.1 EDV48995.1 CM000160 EDW95637.1 GFDL01007911 JAV27134.1 CP012526 ALC47792.1 GDHF01000894 JAI51420.1 CM000070 EAL28768.2 GDHF01030580 JAI21734.1 GAKP01015995 JAC42957.1 JXUM01044209 JXUM01044210 KQ561391 KXJ78725.1 J03154 BT009978 GAKP01022176 JAC36776.1 AAZX01002213 UFQT01000091 SSX19534.1 GDHF01015149 JAI37165.1 CH902623 EDV30269.1 CH479185 EDW37931.1 CH940656 EDW58468.1 GBXI01007200 JAD07092.1 EDX12840.1 CH933806 EDW16999.2 GL453666 EFN75742.1 JXJN01008576 EZ422577 ADD18853.1 CH480815 EDW42071.1 CCAG010004727 QOIP01000012 RLU16063.1 KK107185 EZA55838.1 AY672784 AAV91006.1 EDW97471.1 GAKP01023416 JAC35542.1 GBXI01012101 JAD02191.1 GEZM01061363 JAV70514.1 GEZM01061360 JAV70519.1 GEZM01097973 JAV54026.1 OUUW01000013 SPP87780.1 GBYB01001065 JAG70832.1 CH964232 EDW81243.1 APCN01002556 AAAB01008880 EAA08507.4 GFDL01009899 JAV25146.1
PCG76208.1 KY680241 ATD13322.1 AY677082 AAW24481.1 KT751522 AML39475.1 BABH01025543 BABH01025544 AK401411 BAM18033.1 KT751521 AML39474.1 GAIX01000374 JAA92186.1 KQ460936 KPJ10700.1 KQ459547 KPJ00023.1 AGBW02012316 OWR45231.1 AB009670 BABH01025554 BAA76308.1 JQ581598 AFX82594.1 AGBW02009696 OWR50099.1 KT006138 ANH58163.1 DQ115323 AAZ91696.1 AY077643 AAL76085.1 AB264091 BAF43530.1 AK405456 BAM20768.1 AY672790 AAV91012.1 AAZ91695.1 DQ115324 AAZ91697.1 AAZ91694.1 AY672788 AAV91010.1 JTDY01003981 KOB68749.1 AE014297 AF145611 BT125977 AAD38586.1 ADX35956.1 KT006137 ANH58162.1 GU229936 ADA84072.3 GAMC01000094 JAC06462.1 CH954181 EDV47759.1 OUUW01000014 SPP88304.1 SPP88303.1 KQ434809 KZC06390.1 CVRI01000070 CRL07282.1 GEZM01066955 JAV67623.1 GALX01002154 JAB66312.1 KZ288322 PBC28172.1 CM000364 EDX11579.1 DS232039 EDS32818.1 BT127992 AEE62954.1 GAKP01002127 JAC56825.1 GBXI01016739 GBXI01006351 JAC97552.1 JAD07941.1 CH477325 EAT43491.1 JXUM01042955 JXUM01042956 KQ561344 KXJ78893.1 GAPW01002093 JAC11505.1 EDV48995.1 CM000160 EDW95637.1 GFDL01007911 JAV27134.1 CP012526 ALC47792.1 GDHF01000894 JAI51420.1 CM000070 EAL28768.2 GDHF01030580 JAI21734.1 GAKP01015995 JAC42957.1 JXUM01044209 JXUM01044210 KQ561391 KXJ78725.1 J03154 BT009978 GAKP01022176 JAC36776.1 AAZX01002213 UFQT01000091 SSX19534.1 GDHF01015149 JAI37165.1 CH902623 EDV30269.1 CH479185 EDW37931.1 CH940656 EDW58468.1 GBXI01007200 JAD07092.1 EDX12840.1 CH933806 EDW16999.2 GL453666 EFN75742.1 JXJN01008576 EZ422577 ADD18853.1 CH480815 EDW42071.1 CCAG010004727 QOIP01000012 RLU16063.1 KK107185 EZA55838.1 AY672784 AAV91006.1 EDW97471.1 GAKP01023416 JAC35542.1 GBXI01012101 JAD02191.1 GEZM01061363 JAV70514.1 GEZM01061360 JAV70519.1 GEZM01097973 JAV54026.1 OUUW01000013 SPP87780.1 GBYB01001065 JAG70832.1 CH964232 EDW81243.1 APCN01002556 AAAB01008880 EAA08507.4 GFDL01009899 JAV25146.1
Proteomes
UP000218220
UP000005204
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000000803 UP000008711 UP000268350 UP000076502 UP000192221 UP000183832 UP000242457 UP000000304 UP000002320 UP000095300 UP000095301 UP000008820 UP000069940 UP000249989 UP000002282 UP000092553 UP000001819 UP000002358 UP000007801 UP000008744 UP000008792 UP000075901 UP000009192 UP000008237 UP000092460 UP000001292 UP000092444 UP000279307 UP000053097 UP000092443 UP000078200 UP000007798 UP000075840 UP000007062
UP000000803 UP000008711 UP000268350 UP000076502 UP000192221 UP000183832 UP000242457 UP000000304 UP000002320 UP000095300 UP000095301 UP000008820 UP000069940 UP000249989 UP000002282 UP000092553 UP000001819 UP000002358 UP000007801 UP000008744 UP000008792 UP000075901 UP000009192 UP000008237 UP000092460 UP000001292 UP000092444 UP000279307 UP000053097 UP000092443 UP000078200 UP000007798 UP000075840 UP000007062
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
Q6Y2X4
Q5DI98
A0A2A4JWX6
A0A290U627
Q49QW0
A0A126U9N5
+ More
H9JW83 I4DJE3 A0A140EQ25 S4PHS5 A0A0N1INY1 A0A0N1IMQ8 A0A212EUU4 Q9XXV0 A0A0U1WT40 A0A212F8R3 A0A173GPF0 Q2FAY5 Q8I917 A1IIA5 I4DS78 Q5MPC0 Q2FAY6 Q2FAY4 Q2FAY7 Q5MPC2 A0A0L7L0F2 A0A126GUP6-3 A0A126GUP6-2 A0A173GPF7 A0A126GUP6 D2XPX4 W8CBZ2 B3P1Y6 A0A3B0K246 A0A3B0K4S8 A0A154P5I7 A0A1W4VLE8 A0A1J1J5N3 A0A1Y1L1S6 V5GX21 A0A2A3EA55 B4QUZ8 B0WQE7 J3JXM3 A0A034WRL1 A0A1I8PP27 A0A1I8NE97 A0A0A1WGL4 Q17B76 A0A182G580 A0A023EQV6 B3P0U7 B4PTQ4 A0A1Q3FHX1 A0A0M4EWB3 A0A0K8WKM9 Q295E4 A0A1I8N0W4 A0A0K8U524 A0A034VLF9 A0A182G5Z0 P13582 A0A034V0K7 K7J980 A0A336LMT0 A0A0K8VF53 B3MSM9 B4GM57 B4MBJ7 A0A1W4VIJ6 A0A0A1X7H3 A0A182SX76 B4QYJ5 B4KCX7 E2C8L2 A0A1B0B4Z6 D3TMC5 B4HDX9 A0A1B0FFU3 A0A3L8D7J0 A0A026WIN7 Q5MPC6 A0A1A9YQ00 B4PS51 A0A034V075 A0A0A1WU83 A0A1Y1LA41 A0A1A9UFJ1 A0A1Y1LA48 A0A1Y1K3W7 A0A3B0KH63 A0A0C9QZC0 B4N7Z2 A0A182IIR9 Q7QB73 A0A2Y9D4S9 A0A1Q3FC43
H9JW83 I4DJE3 A0A140EQ25 S4PHS5 A0A0N1INY1 A0A0N1IMQ8 A0A212EUU4 Q9XXV0 A0A0U1WT40 A0A212F8R3 A0A173GPF0 Q2FAY5 Q8I917 A1IIA5 I4DS78 Q5MPC0 Q2FAY6 Q2FAY4 Q2FAY7 Q5MPC2 A0A0L7L0F2 A0A126GUP6-3 A0A126GUP6-2 A0A173GPF7 A0A126GUP6 D2XPX4 W8CBZ2 B3P1Y6 A0A3B0K246 A0A3B0K4S8 A0A154P5I7 A0A1W4VLE8 A0A1J1J5N3 A0A1Y1L1S6 V5GX21 A0A2A3EA55 B4QUZ8 B0WQE7 J3JXM3 A0A034WRL1 A0A1I8PP27 A0A1I8NE97 A0A0A1WGL4 Q17B76 A0A182G580 A0A023EQV6 B3P0U7 B4PTQ4 A0A1Q3FHX1 A0A0M4EWB3 A0A0K8WKM9 Q295E4 A0A1I8N0W4 A0A0K8U524 A0A034VLF9 A0A182G5Z0 P13582 A0A034V0K7 K7J980 A0A336LMT0 A0A0K8VF53 B3MSM9 B4GM57 B4MBJ7 A0A1W4VIJ6 A0A0A1X7H3 A0A182SX76 B4QYJ5 B4KCX7 E2C8L2 A0A1B0B4Z6 D3TMC5 B4HDX9 A0A1B0FFU3 A0A3L8D7J0 A0A026WIN7 Q5MPC6 A0A1A9YQ00 B4PS51 A0A034V075 A0A0A1WU83 A0A1Y1LA41 A0A1A9UFJ1 A0A1Y1LA48 A0A1Y1K3W7 A0A3B0KH63 A0A0C9QZC0 B4N7Z2 A0A182IIR9 Q7QB73 A0A2Y9D4S9 A0A1Q3FC43
PDB
2OLG
E-value=2.4764e-45,
Score=459
Ontologies
GO
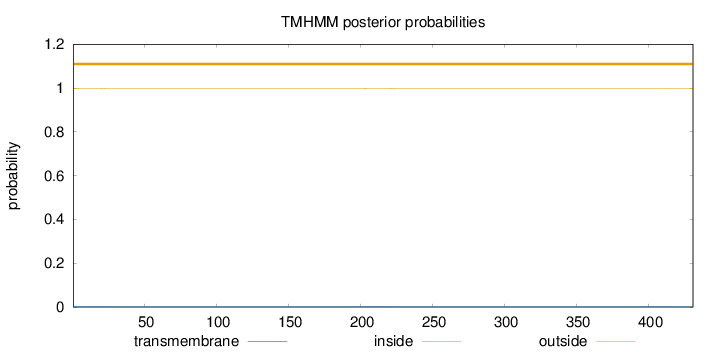
Topology
Subcellular location
Secreted
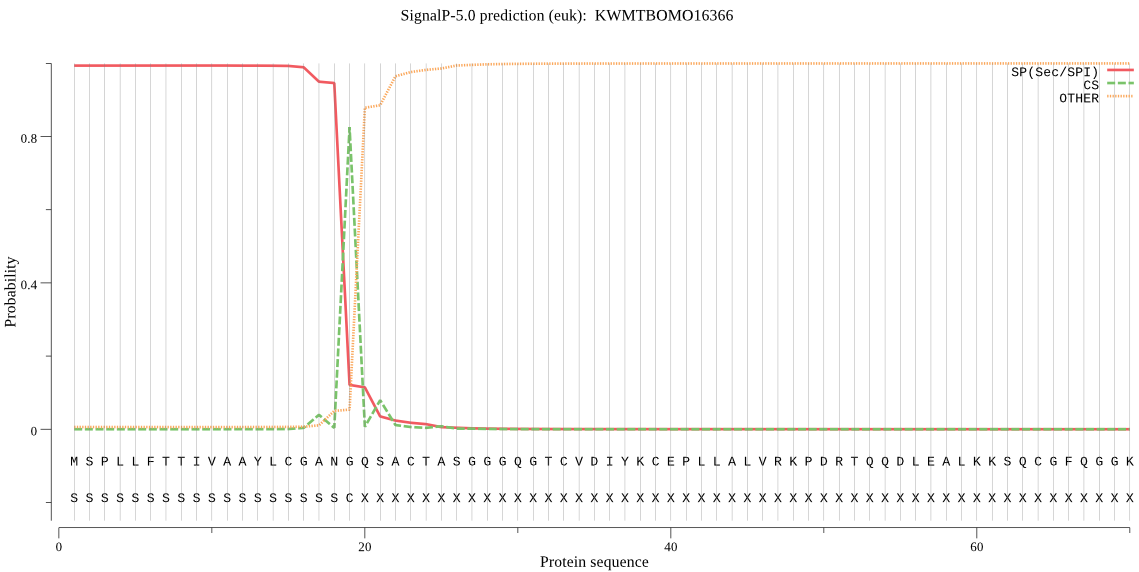
SignalP
Position: 1 - 19,
Likelihood: 0.993707
Length:
431
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0637500000000001
Exp number, first 60 AAs:
0.02197
Total prob of N-in:
0.00245
outside
1 - 431
Population Genetic Test Statistics
Pi
244.464067
Theta
159.471328
Tajima's D
1.45855
CLR
0.052049
CSRT
0.779211039448028
Interpretation
Uncertain
