Gene
KWMTBOMO16362
Annotation
hemolymph_proteinase_10_[Manduca_sexta]
Full name
CLIP domain-containing serine protease 2
+ More
Serine protease easter
Serine protease easter
Alternative Name
BzArgOEtase
Location in the cell
Extracellular Reliability : 2.401
Sequence
CDS
ATGTATAGTATAAAATTATATTTGTGCATCGCGGCGATTTGCGTATTTGTGTTTCTTGAATTCCCAAAATTTAAGGGAAATGAGCACAGATGTGGAATTGAAGCCAGCGGCAACCTTATCCATCACAACCCCTGGTTAGTCTACTTGGAGTACTACGTGGGCGATCATCAGAAGGAATACAGGTGTTCCGGCACGCTGATCAGCCCTCGGCACGTCGTAACAGCAGCTCATTGTGTCAGGAAAATAAAGTTCACCCGTCTGGCGGCTCGTCTCGGCGACTATGACGTTTCAACGGAAAAGGACTGCGTGACGGGCTTGTGCTCTGATCCTAACCTATCTATCCAAGTTTCGGAGATAATCGTCCACCCCGACTACGATGGAAAAGAAGGGGATATCGCGATTCTGAAACTGGACACGGACGCGCCATATACAGACTTCATTCGGCCGATCTGCCTTCCATCCAATCCGATTAAGGGCAACGTCACCCTTTACGCCACCGGCTGGGGGGAGATCCCCACTATAGGAATGTACAGTAATATCAAGAAGATCCTGCCATTACCCAACTGGTCTCTAGAGCAATGCCATAGAGTCTTCGAGAATCTGCCAGATAAGGTGCTCTGTGCTGGAGGCGAAGAAGGCGTTGATACTTGCCGCGGAGACTCTGGTGGTACTCTTGCTCTGGTGAAGGAGAGAGTGGAGTTCGTCGGAGTCACCAGCACTGGGTGCCAGCGATGCGGAACCAAGGGCTACCCAGCTGTATACGTTAGTGTCTATGACTACATAGATTGGATAAGCTTGGTCACGAAACACTGA
Protein
MYSIKLYLCIAAICVFVFLEFPKFKGNEHRCGIEASGNLIHHNPWLVYLEYYVGDHQKEYRCSGTLISPRHVVTAAHCVRKIKFTRLAARLGDYDVSTEKDCVTGLCSDPNLSIQVSEIIVHPDYDGKEGDIAILKLDTDAPYTDFIRPICLPSNPIKGNVTLYATGWGEIPTIGMYSNIKKILPLPNWSLEQCHRVFENLPDKVLCAGGEEGVDTCRGDSGGTLALVKERVEFVGVTSTGCQRCGTKGYPAVYVSVYDYIDWISLVTKH
Summary
Description
Endopeptidase with selective post-Arg cleavage site (PubMed:16399077, PubMed:7737188). Might play a role in innate immunity in response to fungal and Gram-positive bacterial infections (PubMed:16399077).
Component of the extracellular signaling pathway that establishes the dorsal-ventral pathway of the embryo (PubMed:9477324, PubMed:2107028, PubMed:12493753). Three proteases; ndl, gd and snk process easter to create active easter (PubMed:9477324). Active easter defines cell identities along the dorsal-ventral continuum by activating the spz ligand for the Tl receptor in the ventral region of the embryo (PubMed:9477324).
Component of the extracellular signaling pathway that establishes the dorsal-ventral pathway of the embryo (PubMed:9477324, PubMed:2107028, PubMed:12493753). Three proteases; ndl, gd and snk process easter to create active easter (PubMed:9477324). Active easter defines cell identities along the dorsal-ventral continuum by activating the spz ligand for the Tl receptor in the ventral region of the embryo (PubMed:9477324).
Subunit
In the active form, heterodimer of a light chain and a heavy chain; disulfide-linked.
Similarity
Belongs to the peptidase S1 family.
Belongs to the peptidase S1 family. CLIP subfamily.
Belongs to the peptidase S1 family. CLIP subfamily.
Keywords
Calcium
Cleavage on pair of basic residues
Complete proteome
Direct protein sequencing
Disulfide bond
Glycoprotein
Hydrolase
Metal-binding
Protease
Reference proteome
Secreted
Serine protease
Signal
Zymogen
Developmental protein
Feature
chain CLIP domain-containing serine protease 2 light chain
propeptide Activation peptide
propeptide Activation peptide
Uniprot
A0A2H1WEH9
A0A2A4JXI6
Q5MPC4
A0A2H1W881
Q5MPC7
A0A2A4JQ35
+ More
A0A3S2P3X8 Q5MGE4 A0A0T6AZ73 B4M4J5 A0A2M4DQK3 A0A1I8N0W4 W5J9Y3 I4DS78 A0A182S527 A0A2H1WUN7 A0A3F2YTZ8 A0A0L0CE89 A0A1J1J5N3 A0A1I8PP27 A0A212F949 A0A0B4KGQ4 Q7QKL2 A0A1I8JTB4 A0A023EQB0 A0A2C9GV95 A0A2M3Z9P7 A0A2M3ZA33 A0A182GNZ3 B4K6S5 D6W9Z5 B3MSM9 A0A139WN30 A0A2A4JWX6 Q9Y1K6 A0A1W7R8M0 T1DQX5 D2XPX4 A0A0L0C6E5 A0A182YKD5 E2C8L2 A0A212FEW0 A0A290U627 A0A1Q3FEM6 A0A2M4AJX7 A0A2M4BSD0 A0A2M4BSA3 A0A2M4BS12 A0A1S4F560 B3P0U7 A0A3F2YU16 A0A2M4AJZ4 A0A2C9GV93 A0A2C9GWT4 A0A182VAF6 A0A3F2YTX9 B4KCX7 I4DJE3 A0A084VUH0 Q2VG86 A0A182T262 A0A2C9GW66 Q49QW0 B4QYJ5 B0WA88 B4HDX9 A0A1W4VIJ6 C6ZQY3 A0A2M4BRQ8 Q17EX4 P13582 A0A182R026 A0A182QL12 Q17EX5 U5EHU2 A0A2M4BS19 B4PS51 A0A2M4BS35 A0A2M4BS37 W5J6E1 B5DWB5 A0A2M4AR21 W8BLL3 A0A182LCK2 A0A0N1INY1 T1DU92 A1IIA5 A0A1I8PZ22 Q294N1 B4GMV5 A0A1B0G9G5 A0A182XUS5 A0A2C9GTT7 A0A182XUW4 Q17B76 B4JS10 B4N7Z2 Q7PQR9 T1P924 A0A0A1WGL4
A0A3S2P3X8 Q5MGE4 A0A0T6AZ73 B4M4J5 A0A2M4DQK3 A0A1I8N0W4 W5J9Y3 I4DS78 A0A182S527 A0A2H1WUN7 A0A3F2YTZ8 A0A0L0CE89 A0A1J1J5N3 A0A1I8PP27 A0A212F949 A0A0B4KGQ4 Q7QKL2 A0A1I8JTB4 A0A023EQB0 A0A2C9GV95 A0A2M3Z9P7 A0A2M3ZA33 A0A182GNZ3 B4K6S5 D6W9Z5 B3MSM9 A0A139WN30 A0A2A4JWX6 Q9Y1K6 A0A1W7R8M0 T1DQX5 D2XPX4 A0A0L0C6E5 A0A182YKD5 E2C8L2 A0A212FEW0 A0A290U627 A0A1Q3FEM6 A0A2M4AJX7 A0A2M4BSD0 A0A2M4BSA3 A0A2M4BS12 A0A1S4F560 B3P0U7 A0A3F2YU16 A0A2M4AJZ4 A0A2C9GV93 A0A2C9GWT4 A0A182VAF6 A0A3F2YTX9 B4KCX7 I4DJE3 A0A084VUH0 Q2VG86 A0A182T262 A0A2C9GW66 Q49QW0 B4QYJ5 B0WA88 B4HDX9 A0A1W4VIJ6 C6ZQY3 A0A2M4BRQ8 Q17EX4 P13582 A0A182R026 A0A182QL12 Q17EX5 U5EHU2 A0A2M4BS19 B4PS51 A0A2M4BS35 A0A2M4BS37 W5J6E1 B5DWB5 A0A2M4AR21 W8BLL3 A0A182LCK2 A0A0N1INY1 T1DU92 A1IIA5 A0A1I8PZ22 Q294N1 B4GMV5 A0A1B0G9G5 A0A182XUS5 A0A2C9GTT7 A0A182XUW4 Q17B76 B4JS10 B4N7Z2 Q7PQR9 T1P924 A0A0A1WGL4
EC Number
3.4.21.-
Pubmed
15944088
16023793
17994087
25315136
20920257
23761445
+ More
22651552 26108605 22118469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12364791 14747013 17210077 24945155 26483478 18362917 19820115 10646969 25244985 20798317 28953952 17510324 24438588 16399077 19121390 7737188 20496585 2492450 9477324 9533958 2107028 12493753 17550304 15632085 24495485 20966253 26354079 17307003 25830018
22651552 26108605 22118469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12364791 14747013 17210077 24945155 26483478 18362917 19820115 10646969 25244985 20798317 28953952 17510324 24438588 16399077 19121390 7737188 20496585 2492450 9477324 9533958 2107028 12493753 17550304 15632085 24495485 20966253 26354079 17307003 25830018
EMBL
ODYU01008137
SOQ51490.1
NWSH01000467
PCG76213.1
AY672786
AAV91008.1
+ More
ODYU01006975 SOQ49305.1 AY672783 AAV91005.1 NWSH01000925 PCG73522.1 RSAL01000029 RVE51737.1 AY829842 AAV91456.1 LJIG01022469 KRT80412.1 CH940652 EDW59556.1 GGFL01015657 MBW79835.1 ADMH02002074 ETN59569.1 AK405456 BAM20768.1 ODYU01011137 SOQ56686.1 JRES01000503 KNC30561.1 CVRI01000070 CRL07282.1 AGBW02009640 OWR50264.1 AE014297 AGB95978.1 AAAB01008794 EAA03566.2 APCN01002147 GAPW01001836 JAC11762.1 GGFM01004490 MBW25241.1 GGFM01004547 MBW25298.1 JXUM01015012 JXUM01015013 KQ560420 KXJ82504.1 CH933806 EDW14191.1 KQ971312 EEZ99203.2 CH902623 EDV30269.1 KYB29266.1 PCG76208.1 AF117749 AAD38335.1 GEHC01000168 JAV47477.1 GAMD01000905 JAB00686.1 GU229936 ADA84072.3 JRES01000835 KNC27963.1 GL453666 EFN75742.1 AGBW02008891 OWR52286.1 KY680241 ATD13322.1 GFDL01009047 JAV25998.1 GGFK01007768 MBW41089.1 GGFJ01006752 MBW55893.1 GGFJ01006751 MBW55892.1 GGFJ01006726 MBW55867.1 CH954181 EDV48995.1 GGFK01007753 MBW41074.1 EDW16999.2 AK401411 BAM18033.1 ATLV01016791 KE525113 KFB41614.1 AB035418 AY061936 DQ157441 BABH01019990 AAL31707.1 ABB58762.1 BAE73254.1 AXCN02000165 AY677082 AAW24481.1 CM000364 EDX12840.1 DS231869 EDS40917.1 CH480815 EDW42071.1 EZ114934 ACU30987.1 GGFJ01006522 MBW55663.1 CH477278 EAT45085.1 J03154 BT009978 EAT45084.1 GANO01002884 JAB56987.1 GGFJ01006671 MBW55812.1 CM000160 EDW97471.1 GGFJ01006728 MBW55869.1 GGFJ01006754 MBW55895.1 ETN59571.1 CM000070 EDY68533.1 GGFK01009924 MBW43245.1 GAMC01016016 JAB90539.1 KQ460936 KPJ10700.1 GAMD01000359 JAB01232.1 AB264091 BAF43530.1 EAL28934.1 CH479185 EDW38179.1 CCAG010018107 CH477325 EAT43491.1 CH916373 EDV94550.1 CH964232 EDW81243.1 AAAB01008879 EAA08404.4 KA645202 AFP59831.1 GBXI01016739 GBXI01006351 JAC97552.1 JAD07941.1
ODYU01006975 SOQ49305.1 AY672783 AAV91005.1 NWSH01000925 PCG73522.1 RSAL01000029 RVE51737.1 AY829842 AAV91456.1 LJIG01022469 KRT80412.1 CH940652 EDW59556.1 GGFL01015657 MBW79835.1 ADMH02002074 ETN59569.1 AK405456 BAM20768.1 ODYU01011137 SOQ56686.1 JRES01000503 KNC30561.1 CVRI01000070 CRL07282.1 AGBW02009640 OWR50264.1 AE014297 AGB95978.1 AAAB01008794 EAA03566.2 APCN01002147 GAPW01001836 JAC11762.1 GGFM01004490 MBW25241.1 GGFM01004547 MBW25298.1 JXUM01015012 JXUM01015013 KQ560420 KXJ82504.1 CH933806 EDW14191.1 KQ971312 EEZ99203.2 CH902623 EDV30269.1 KYB29266.1 PCG76208.1 AF117749 AAD38335.1 GEHC01000168 JAV47477.1 GAMD01000905 JAB00686.1 GU229936 ADA84072.3 JRES01000835 KNC27963.1 GL453666 EFN75742.1 AGBW02008891 OWR52286.1 KY680241 ATD13322.1 GFDL01009047 JAV25998.1 GGFK01007768 MBW41089.1 GGFJ01006752 MBW55893.1 GGFJ01006751 MBW55892.1 GGFJ01006726 MBW55867.1 CH954181 EDV48995.1 GGFK01007753 MBW41074.1 EDW16999.2 AK401411 BAM18033.1 ATLV01016791 KE525113 KFB41614.1 AB035418 AY061936 DQ157441 BABH01019990 AAL31707.1 ABB58762.1 BAE73254.1 AXCN02000165 AY677082 AAW24481.1 CM000364 EDX12840.1 DS231869 EDS40917.1 CH480815 EDW42071.1 EZ114934 ACU30987.1 GGFJ01006522 MBW55663.1 CH477278 EAT45085.1 J03154 BT009978 EAT45084.1 GANO01002884 JAB56987.1 GGFJ01006671 MBW55812.1 CM000160 EDW97471.1 GGFJ01006728 MBW55869.1 GGFJ01006754 MBW55895.1 ETN59571.1 CM000070 EDY68533.1 GGFK01009924 MBW43245.1 GAMC01016016 JAB90539.1 KQ460936 KPJ10700.1 GAMD01000359 JAB01232.1 AB264091 BAF43530.1 EAL28934.1 CH479185 EDW38179.1 CCAG010018107 CH477325 EAT43491.1 CH916373 EDV94550.1 CH964232 EDW81243.1 AAAB01008879 EAA08404.4 KA645202 AFP59831.1 GBXI01016739 GBXI01006351 JAC97552.1 JAD07941.1
Proteomes
UP000218220
UP000283053
UP000008792
UP000095301
UP000000673
UP000075900
+ More
UP000075881 UP000037069 UP000183832 UP000095300 UP000007151 UP000000803 UP000007062 UP000075840 UP000075884 UP000069940 UP000249989 UP000009192 UP000007266 UP000007801 UP000076408 UP000008237 UP000008711 UP000075903 UP000030765 UP000005204 UP000075901 UP000075886 UP000000304 UP000002320 UP000001292 UP000192221 UP000008820 UP000002282 UP000001819 UP000075882 UP000053240 UP000008744 UP000092444 UP000076407 UP000001070 UP000007798
UP000075881 UP000037069 UP000183832 UP000095300 UP000007151 UP000000803 UP000007062 UP000075840 UP000075884 UP000069940 UP000249989 UP000009192 UP000007266 UP000007801 UP000076408 UP000008237 UP000008711 UP000075903 UP000030765 UP000005204 UP000075901 UP000075886 UP000000304 UP000002320 UP000001292 UP000192221 UP000008820 UP000002282 UP000001819 UP000075882 UP000053240 UP000008744 UP000092444 UP000076407 UP000001070 UP000007798
Interpro
SUPFAM
SSF50494
SSF50494
Gene 3D
CDD
ProteinModelPortal
A0A2H1WEH9
A0A2A4JXI6
Q5MPC4
A0A2H1W881
Q5MPC7
A0A2A4JQ35
+ More
A0A3S2P3X8 Q5MGE4 A0A0T6AZ73 B4M4J5 A0A2M4DQK3 A0A1I8N0W4 W5J9Y3 I4DS78 A0A182S527 A0A2H1WUN7 A0A3F2YTZ8 A0A0L0CE89 A0A1J1J5N3 A0A1I8PP27 A0A212F949 A0A0B4KGQ4 Q7QKL2 A0A1I8JTB4 A0A023EQB0 A0A2C9GV95 A0A2M3Z9P7 A0A2M3ZA33 A0A182GNZ3 B4K6S5 D6W9Z5 B3MSM9 A0A139WN30 A0A2A4JWX6 Q9Y1K6 A0A1W7R8M0 T1DQX5 D2XPX4 A0A0L0C6E5 A0A182YKD5 E2C8L2 A0A212FEW0 A0A290U627 A0A1Q3FEM6 A0A2M4AJX7 A0A2M4BSD0 A0A2M4BSA3 A0A2M4BS12 A0A1S4F560 B3P0U7 A0A3F2YU16 A0A2M4AJZ4 A0A2C9GV93 A0A2C9GWT4 A0A182VAF6 A0A3F2YTX9 B4KCX7 I4DJE3 A0A084VUH0 Q2VG86 A0A182T262 A0A2C9GW66 Q49QW0 B4QYJ5 B0WA88 B4HDX9 A0A1W4VIJ6 C6ZQY3 A0A2M4BRQ8 Q17EX4 P13582 A0A182R026 A0A182QL12 Q17EX5 U5EHU2 A0A2M4BS19 B4PS51 A0A2M4BS35 A0A2M4BS37 W5J6E1 B5DWB5 A0A2M4AR21 W8BLL3 A0A182LCK2 A0A0N1INY1 T1DU92 A1IIA5 A0A1I8PZ22 Q294N1 B4GMV5 A0A1B0G9G5 A0A182XUS5 A0A2C9GTT7 A0A182XUW4 Q17B76 B4JS10 B4N7Z2 Q7PQR9 T1P924 A0A0A1WGL4
A0A3S2P3X8 Q5MGE4 A0A0T6AZ73 B4M4J5 A0A2M4DQK3 A0A1I8N0W4 W5J9Y3 I4DS78 A0A182S527 A0A2H1WUN7 A0A3F2YTZ8 A0A0L0CE89 A0A1J1J5N3 A0A1I8PP27 A0A212F949 A0A0B4KGQ4 Q7QKL2 A0A1I8JTB4 A0A023EQB0 A0A2C9GV95 A0A2M3Z9P7 A0A2M3ZA33 A0A182GNZ3 B4K6S5 D6W9Z5 B3MSM9 A0A139WN30 A0A2A4JWX6 Q9Y1K6 A0A1W7R8M0 T1DQX5 D2XPX4 A0A0L0C6E5 A0A182YKD5 E2C8L2 A0A212FEW0 A0A290U627 A0A1Q3FEM6 A0A2M4AJX7 A0A2M4BSD0 A0A2M4BSA3 A0A2M4BS12 A0A1S4F560 B3P0U7 A0A3F2YU16 A0A2M4AJZ4 A0A2C9GV93 A0A2C9GWT4 A0A182VAF6 A0A3F2YTX9 B4KCX7 I4DJE3 A0A084VUH0 Q2VG86 A0A182T262 A0A2C9GW66 Q49QW0 B4QYJ5 B0WA88 B4HDX9 A0A1W4VIJ6 C6ZQY3 A0A2M4BRQ8 Q17EX4 P13582 A0A182R026 A0A182QL12 Q17EX5 U5EHU2 A0A2M4BS19 B4PS51 A0A2M4BS35 A0A2M4BS37 W5J6E1 B5DWB5 A0A2M4AR21 W8BLL3 A0A182LCK2 A0A0N1INY1 T1DU92 A1IIA5 A0A1I8PZ22 Q294N1 B4GMV5 A0A1B0G9G5 A0A182XUS5 A0A2C9GTT7 A0A182XUW4 Q17B76 B4JS10 B4N7Z2 Q7PQR9 T1P924 A0A0A1WGL4
PDB
2OLG
E-value=2.23805e-35,
Score=371
Ontologies
GO
Topology
Subcellular location
Secreted
Secreted in the hemolymph. With evidence from 6 publications.
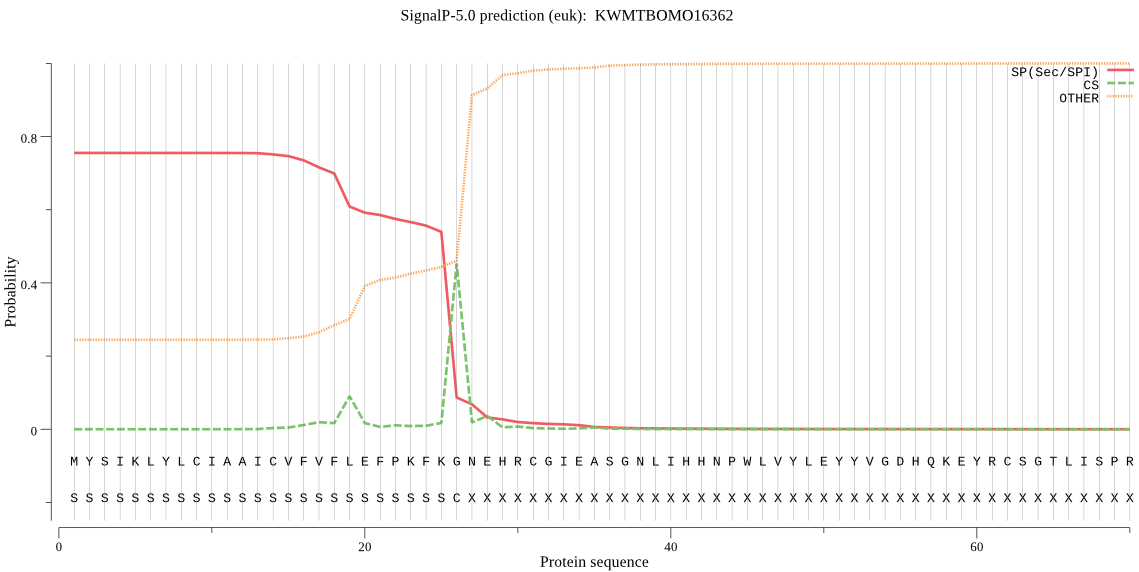
SignalP
Position: 1 - 26,
Likelihood: 0.755356
Length:
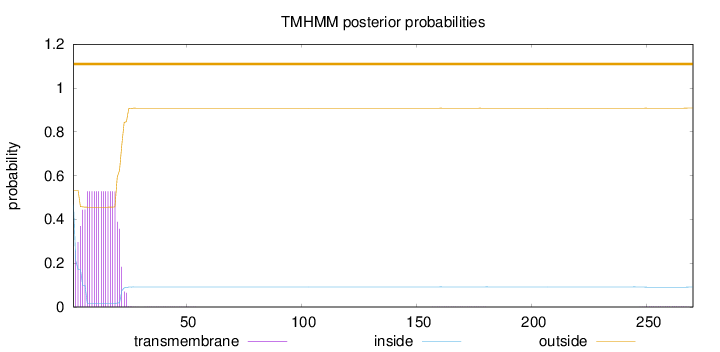
270
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.79067999999998
Exp number, first 60 AAs:
9.75252000000001
Total prob of N-in:
0.46817
outside
1 - 270
Population Genetic Test Statistics
Pi
32.855533
Theta
13.917719
Tajima's D
-1.234908
CLR
39.599476
CSRT
0.0983450827458627
Interpretation
Uncertain
