Gene
KWMTBOMO16361
Pre Gene Modal
BGIBMGA013751
Annotation
PREDICTED:_serine_protease_easter-like_[Bombyx_mori]
Full name
CLIP domain-containing serine protease 2
Alternative Name
BzArgOEtase
Location in the cell
Extracellular Reliability : 2.228
Sequence
CDS
ATGAGATATGTCTGCCTGTTAATTGTTAAGTTAATGATATCCTCTTGGAGACTAATACACCAAGACCCCTGGCTGGTTCTGCTCGAGTACCACCGTGAAGGAAGACGCACAGGTGCCAGGTGTGGAGGTACGCTGGTCGGACCCCGTCACGTCATCACTGCTGCTCACTGCGTCAGGAACGCCTTCTTGGATCGGTTAACCGTACGCTTGGGGGAATACGATACGGCCTCTAAAATTGACTGCTCTGAAGGTGTTTGTTCTGACAAAGTTGTTCGTATCAATGTCACCGAGATCTTCGCGCATCCAGACTACGTTGATAAACAAAATGACATCGCTGTGTTGAAGCTAGAGAAGGATGCTCCTTATACTGATTTCATCCGTCCTGTTTGTCTTCCTACTGGAACGTTAAGTCCTAGGACATTGTTCTTGGCCTCAGGATGGGGACAGCAAGCAAACGGCTTATATAGTCACATCAAAAAGCTGATACCATTACCGAACTGGTCGATTGAAGATTGCCAAGCAATGTACAAGCAAACAAAGTTGCCCGATAAAATCATTTGTGCTGGTGGCGAAAGAGGAATGGACACGTGCCGCGGGGATTCCGGTGGGCCATTGGTGGTCATGAAGAGGAGATCTGAGTTGTGGGGCATCACCAGCGCCGGCCACGTCCATTGCGGTTCACAAGGATATCCCGGGATTTATACGAATGTTACCATGTTCTTGGACTGGATCGGAGATGTAATGAAGATTTGA
Protein
MRYVCLLIVKLMISSWRLIHQDPWLVLLEYHREGRRTGARCGGTLVGPRHVITAAHCVRNAFLDRLTVRLGEYDTASKIDCSEGVCSDKVVRINVTEIFAHPDYVDKQNDIAVLKLEKDAPYTDFIRPVCLPTGTLSPRTLFLASGWGQQANGLYSHIKKLIPLPNWSIEDCQAMYKQTKLPDKIICAGGERGMDTCRGDSGGPLVVMKRRSELWGITSAGHVHCGSQGYPGIYTNVTMFLDWIGDVMKI
Summary
Description
Endopeptidase with selective post-Arg cleavage site (PubMed:16399077, PubMed:7737188). Might play a role in innate immunity in response to fungal and Gram-positive bacterial infections (PubMed:16399077).
Subunit
In the active form, heterodimer of a light chain and a heavy chain; disulfide-linked.
Similarity
Belongs to the peptidase S1 family.
Belongs to the peptidase S1 family. CLIP subfamily.
Belongs to the peptidase S1 family. CLIP subfamily.
Keywords
Calcium
Cleavage on pair of basic residues
Complete proteome
Direct protein sequencing
Disulfide bond
Glycoprotein
Hydrolase
Metal-binding
Protease
Reference proteome
Secreted
Serine protease
Signal
Zymogen
Feature
chain CLIP domain-containing serine protease 2 light chain
Uniprot
Q5MPC4
A0A2H1WEH9
A0A2H1W881
A0A2A4JQ35
A0A2A4JXI6
A0A3S2P3X8
+ More
Q5MPC7 Q5MGE4 A0A1I8PP27 A0A1J1J5N3 A0A139WN30 A0A2H1WUN7 D6W9Z5 A0A3S2M731 A0A2C9GW66 Q17B76 A0A2C9GWT4 A0A1B0DG63 A0A023EPR7 Q2VG86 E2C8L2 A0A0K8UPH0 A0A0K8VIH4 A0A0M9A7E2 A0A023EQV6 A0A1I8N0W4 A0A0L7QXF0 Q49QW0 A0A2A4JWX6 A0A1I8JTB4 A0A026WIN7 A0A212F949 A0A182G5Z0 A0A182G580 Q9Y1K6 A0A084W8U9 A0A023EQB0 A0A0M4F7H7 A0A3L8D7J0 A0A290U615 A0A182GNZ3 A0A1A9UFJ1 Q7QKL2 A0A1A9ZT35 B3F5B1 A0A1Y1KDT6 A0A290U627 A0A173GPF2 A0A2A4JQA1 N6TWW7 A0A212FEW0 A0A0L0CE89 A0A0U1WT79 A0A2A3EA55 A0A182GH79 A0A1W7R8M0 A0A1I8PEC5 A0A1B0B4Z6 A0A1A9YQ00 A0A0M4EWB3 E2AV85 T1P924 B4N925 D6WUF7 A0A151WR74 B4MBJ7 D3TMC5 A0A0C9QZC0 A0A182GNY7 A0A182INH2 A0A182VAF6 A0A1B0FFU3 B1B5K1 A0A2H1VCQ7 S4PKY1 F4WPR9 A0A088A359 A0A2H1VTY9 A0A1I8NE97 A0A154P5I7 A0A1I8PE36 A0A0M4EMT4 A0A2H1V1Q2 A0A0J7NWE6 A0A2C9GVA1 A0A1Y1NG54 A0A3B0KH63 A0A0B4KGQ4 A0A1Y1NG56 A0A1S4F560 B4KCX7 Q49QW1 U4U606 A0A1Y1NL68 B3P1Y6 A0A1A9WC15 A0A195FEH3 A0A158NI84 A0A1Y1NJX5
Q5MPC7 Q5MGE4 A0A1I8PP27 A0A1J1J5N3 A0A139WN30 A0A2H1WUN7 D6W9Z5 A0A3S2M731 A0A2C9GW66 Q17B76 A0A2C9GWT4 A0A1B0DG63 A0A023EPR7 Q2VG86 E2C8L2 A0A0K8UPH0 A0A0K8VIH4 A0A0M9A7E2 A0A023EQV6 A0A1I8N0W4 A0A0L7QXF0 Q49QW0 A0A2A4JWX6 A0A1I8JTB4 A0A026WIN7 A0A212F949 A0A182G5Z0 A0A182G580 Q9Y1K6 A0A084W8U9 A0A023EQB0 A0A0M4F7H7 A0A3L8D7J0 A0A290U615 A0A182GNZ3 A0A1A9UFJ1 Q7QKL2 A0A1A9ZT35 B3F5B1 A0A1Y1KDT6 A0A290U627 A0A173GPF2 A0A2A4JQA1 N6TWW7 A0A212FEW0 A0A0L0CE89 A0A0U1WT79 A0A2A3EA55 A0A182GH79 A0A1W7R8M0 A0A1I8PEC5 A0A1B0B4Z6 A0A1A9YQ00 A0A0M4EWB3 E2AV85 T1P924 B4N925 D6WUF7 A0A151WR74 B4MBJ7 D3TMC5 A0A0C9QZC0 A0A182GNY7 A0A182INH2 A0A182VAF6 A0A1B0FFU3 B1B5K1 A0A2H1VCQ7 S4PKY1 F4WPR9 A0A088A359 A0A2H1VTY9 A0A1I8NE97 A0A154P5I7 A0A1I8PE36 A0A0M4EMT4 A0A2H1V1Q2 A0A0J7NWE6 A0A2C9GVA1 A0A1Y1NG54 A0A3B0KH63 A0A0B4KGQ4 A0A1Y1NG56 A0A1S4F560 B4KCX7 Q49QW1 U4U606 A0A1Y1NL68 B3P1Y6 A0A1A9WC15 A0A195FEH3 A0A158NI84 A0A1Y1NJX5
EC Number
3.4.21.-
Pubmed
15944088
16023793
18362917
19820115
17510324
24945155
+ More
16399077 19121390 7737188 20798317 25315136 24508170 22118469 26483478 10646969 24438588 30249741 28953952 12364791 14747013 17210077 28004739 23537049 26108605 17994087 20353571 18195005 23622113 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19557749 21347285
16399077 19121390 7737188 20798317 25315136 24508170 22118469 26483478 10646969 24438588 30249741 28953952 12364791 14747013 17210077 28004739 23537049 26108605 17994087 20353571 18195005 23622113 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19557749 21347285
EMBL
AY672786
AAV91008.1
ODYU01008137
SOQ51490.1
ODYU01006975
SOQ49305.1
+ More
NWSH01000925 PCG73522.1 NWSH01000467 PCG76213.1 RSAL01000029 RVE51737.1 AY672783 AAV91005.1 AY829842 AAV91456.1 CVRI01000070 CRL07282.1 KQ971312 KYB29266.1 ODYU01011137 SOQ56686.1 EEZ99203.2 RSAL01000017 RVE52975.1 AXCN02000165 CH477325 EAT43491.1 AJVK01059305 GAPW01002593 JAC11005.1 AB035418 AY061936 DQ157441 BABH01019990 AAL31707.1 ABB58762.1 BAE73254.1 GL453666 EFN75742.1 GDHF01024094 GDHF01019994 JAI28220.1 JAI32320.1 GDHF01013623 JAI38691.1 KQ435719 KOX78755.1 GAPW01002093 JAC11505.1 KQ414704 KOC63285.1 AY677082 AAW24481.1 PCG76208.1 APCN01002147 KK107185 EZA55838.1 AGBW02009640 OWR50264.1 JXUM01044209 JXUM01044210 KQ561391 KXJ78725.1 JXUM01042955 JXUM01042956 KQ561344 KXJ78893.1 AF117749 AAD38335.1 ATLV01021515 KE525319 KFB46643.1 GAPW01001836 JAC11762.1 CP012526 ALC47832.1 QOIP01000012 RLU16063.1 KY680243 ATD13324.1 JXUM01015012 JXUM01015013 KQ560420 KXJ82504.1 AAAB01008794 EAA03566.2 EF614332 ABU98654.1 GEZM01087707 JAV58521.1 KY680241 ATD13322.1 KT006136 ANH58161.1 NWSH01000833 PCG73946.1 APGK01057809 KB741282 ENN70807.1 AGBW02008891 OWR52286.1 JRES01000503 KNC30561.1 JQ581597 AFX82593.1 KZ288322 PBC28172.1 JXUM01063272 KQ562240 KXJ76328.1 GEHC01000168 JAV47477.1 JXJN01008576 ALC47792.1 GL443059 EFN62639.1 KA645202 AFP59831.1 CH964232 EDW81572.2 KQ971352 EFA07559.2 KQ982813 KYQ50307.1 CH940656 EDW58468.1 EZ422577 ADD18853.1 GBYB01001065 JAG70832.1 JXUM01015001 KXJ82498.1 CCAG010004727 AB363980 BAG14262.2 ODYU01001850 SOQ38623.1 GAIX01004295 JAA88265.1 GL888255 EGI63810.1 ODYU01004434 SOQ44299.1 KQ434809 KZC06390.1 ALC45838.1 ODYU01000283 SOQ34775.1 LBMM01001207 KMQ96720.1 GEZM01003115 JAV96914.1 OUUW01000013 SPP87780.1 AE014297 AGB95978.1 GEZM01003116 JAV96913.1 CH933806 EDW16999.2 AY677081 AAW24480.1 KB631684 ERL85365.1 GEZM01003113 JAV96916.1 CH954181 EDV47759.1 KQ981636 KYN38803.1 ADTU01016095 GEZM01003114 JAV96915.1
NWSH01000925 PCG73522.1 NWSH01000467 PCG76213.1 RSAL01000029 RVE51737.1 AY672783 AAV91005.1 AY829842 AAV91456.1 CVRI01000070 CRL07282.1 KQ971312 KYB29266.1 ODYU01011137 SOQ56686.1 EEZ99203.2 RSAL01000017 RVE52975.1 AXCN02000165 CH477325 EAT43491.1 AJVK01059305 GAPW01002593 JAC11005.1 AB035418 AY061936 DQ157441 BABH01019990 AAL31707.1 ABB58762.1 BAE73254.1 GL453666 EFN75742.1 GDHF01024094 GDHF01019994 JAI28220.1 JAI32320.1 GDHF01013623 JAI38691.1 KQ435719 KOX78755.1 GAPW01002093 JAC11505.1 KQ414704 KOC63285.1 AY677082 AAW24481.1 PCG76208.1 APCN01002147 KK107185 EZA55838.1 AGBW02009640 OWR50264.1 JXUM01044209 JXUM01044210 KQ561391 KXJ78725.1 JXUM01042955 JXUM01042956 KQ561344 KXJ78893.1 AF117749 AAD38335.1 ATLV01021515 KE525319 KFB46643.1 GAPW01001836 JAC11762.1 CP012526 ALC47832.1 QOIP01000012 RLU16063.1 KY680243 ATD13324.1 JXUM01015012 JXUM01015013 KQ560420 KXJ82504.1 AAAB01008794 EAA03566.2 EF614332 ABU98654.1 GEZM01087707 JAV58521.1 KY680241 ATD13322.1 KT006136 ANH58161.1 NWSH01000833 PCG73946.1 APGK01057809 KB741282 ENN70807.1 AGBW02008891 OWR52286.1 JRES01000503 KNC30561.1 JQ581597 AFX82593.1 KZ288322 PBC28172.1 JXUM01063272 KQ562240 KXJ76328.1 GEHC01000168 JAV47477.1 JXJN01008576 ALC47792.1 GL443059 EFN62639.1 KA645202 AFP59831.1 CH964232 EDW81572.2 KQ971352 EFA07559.2 KQ982813 KYQ50307.1 CH940656 EDW58468.1 EZ422577 ADD18853.1 GBYB01001065 JAG70832.1 JXUM01015001 KXJ82498.1 CCAG010004727 AB363980 BAG14262.2 ODYU01001850 SOQ38623.1 GAIX01004295 JAA88265.1 GL888255 EGI63810.1 ODYU01004434 SOQ44299.1 KQ434809 KZC06390.1 ALC45838.1 ODYU01000283 SOQ34775.1 LBMM01001207 KMQ96720.1 GEZM01003115 JAV96914.1 OUUW01000013 SPP87780.1 AE014297 AGB95978.1 GEZM01003116 JAV96913.1 CH933806 EDW16999.2 AY677081 AAW24480.1 KB631684 ERL85365.1 GEZM01003113 JAV96916.1 CH954181 EDV47759.1 KQ981636 KYN38803.1 ADTU01016095 GEZM01003114 JAV96915.1
Proteomes
UP000218220
UP000283053
UP000095300
UP000183832
UP000007266
UP000075886
+ More
UP000008820 UP000075900 UP000092462 UP000005204 UP000008237 UP000053105 UP000095301 UP000053825 UP000075840 UP000053097 UP000007151 UP000069940 UP000249989 UP000030765 UP000092553 UP000279307 UP000078200 UP000007062 UP000092445 UP000019118 UP000037069 UP000242457 UP000092460 UP000092443 UP000000311 UP000007798 UP000075809 UP000008792 UP000075880 UP000075903 UP000092444 UP000007755 UP000005203 UP000076502 UP000036403 UP000075884 UP000268350 UP000000803 UP000009192 UP000030742 UP000008711 UP000091820 UP000078541 UP000005205
UP000008820 UP000075900 UP000092462 UP000005204 UP000008237 UP000053105 UP000095301 UP000053825 UP000075840 UP000053097 UP000007151 UP000069940 UP000249989 UP000030765 UP000092553 UP000279307 UP000078200 UP000007062 UP000092445 UP000019118 UP000037069 UP000242457 UP000092460 UP000092443 UP000000311 UP000007798 UP000075809 UP000008792 UP000075880 UP000075903 UP000092444 UP000007755 UP000005203 UP000076502 UP000036403 UP000075884 UP000268350 UP000000803 UP000009192 UP000030742 UP000008711 UP000091820 UP000078541 UP000005205
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
Q5MPC4
A0A2H1WEH9
A0A2H1W881
A0A2A4JQ35
A0A2A4JXI6
A0A3S2P3X8
+ More
Q5MPC7 Q5MGE4 A0A1I8PP27 A0A1J1J5N3 A0A139WN30 A0A2H1WUN7 D6W9Z5 A0A3S2M731 A0A2C9GW66 Q17B76 A0A2C9GWT4 A0A1B0DG63 A0A023EPR7 Q2VG86 E2C8L2 A0A0K8UPH0 A0A0K8VIH4 A0A0M9A7E2 A0A023EQV6 A0A1I8N0W4 A0A0L7QXF0 Q49QW0 A0A2A4JWX6 A0A1I8JTB4 A0A026WIN7 A0A212F949 A0A182G5Z0 A0A182G580 Q9Y1K6 A0A084W8U9 A0A023EQB0 A0A0M4F7H7 A0A3L8D7J0 A0A290U615 A0A182GNZ3 A0A1A9UFJ1 Q7QKL2 A0A1A9ZT35 B3F5B1 A0A1Y1KDT6 A0A290U627 A0A173GPF2 A0A2A4JQA1 N6TWW7 A0A212FEW0 A0A0L0CE89 A0A0U1WT79 A0A2A3EA55 A0A182GH79 A0A1W7R8M0 A0A1I8PEC5 A0A1B0B4Z6 A0A1A9YQ00 A0A0M4EWB3 E2AV85 T1P924 B4N925 D6WUF7 A0A151WR74 B4MBJ7 D3TMC5 A0A0C9QZC0 A0A182GNY7 A0A182INH2 A0A182VAF6 A0A1B0FFU3 B1B5K1 A0A2H1VCQ7 S4PKY1 F4WPR9 A0A088A359 A0A2H1VTY9 A0A1I8NE97 A0A154P5I7 A0A1I8PE36 A0A0M4EMT4 A0A2H1V1Q2 A0A0J7NWE6 A0A2C9GVA1 A0A1Y1NG54 A0A3B0KH63 A0A0B4KGQ4 A0A1Y1NG56 A0A1S4F560 B4KCX7 Q49QW1 U4U606 A0A1Y1NL68 B3P1Y6 A0A1A9WC15 A0A195FEH3 A0A158NI84 A0A1Y1NJX5
Q5MPC7 Q5MGE4 A0A1I8PP27 A0A1J1J5N3 A0A139WN30 A0A2H1WUN7 D6W9Z5 A0A3S2M731 A0A2C9GW66 Q17B76 A0A2C9GWT4 A0A1B0DG63 A0A023EPR7 Q2VG86 E2C8L2 A0A0K8UPH0 A0A0K8VIH4 A0A0M9A7E2 A0A023EQV6 A0A1I8N0W4 A0A0L7QXF0 Q49QW0 A0A2A4JWX6 A0A1I8JTB4 A0A026WIN7 A0A212F949 A0A182G5Z0 A0A182G580 Q9Y1K6 A0A084W8U9 A0A023EQB0 A0A0M4F7H7 A0A3L8D7J0 A0A290U615 A0A182GNZ3 A0A1A9UFJ1 Q7QKL2 A0A1A9ZT35 B3F5B1 A0A1Y1KDT6 A0A290U627 A0A173GPF2 A0A2A4JQA1 N6TWW7 A0A212FEW0 A0A0L0CE89 A0A0U1WT79 A0A2A3EA55 A0A182GH79 A0A1W7R8M0 A0A1I8PEC5 A0A1B0B4Z6 A0A1A9YQ00 A0A0M4EWB3 E2AV85 T1P924 B4N925 D6WUF7 A0A151WR74 B4MBJ7 D3TMC5 A0A0C9QZC0 A0A182GNY7 A0A182INH2 A0A182VAF6 A0A1B0FFU3 B1B5K1 A0A2H1VCQ7 S4PKY1 F4WPR9 A0A088A359 A0A2H1VTY9 A0A1I8NE97 A0A154P5I7 A0A1I8PE36 A0A0M4EMT4 A0A2H1V1Q2 A0A0J7NWE6 A0A2C9GVA1 A0A1Y1NG54 A0A3B0KH63 A0A0B4KGQ4 A0A1Y1NG56 A0A1S4F560 B4KCX7 Q49QW1 U4U606 A0A1Y1NL68 B3P1Y6 A0A1A9WC15 A0A195FEH3 A0A158NI84 A0A1Y1NJX5
PDB
2OLG
E-value=1.51039e-38,
Score=398
Ontologies
GO
Topology
Subcellular location
Secreted
Secreted in the hemolymph. With evidence from 6 publications.
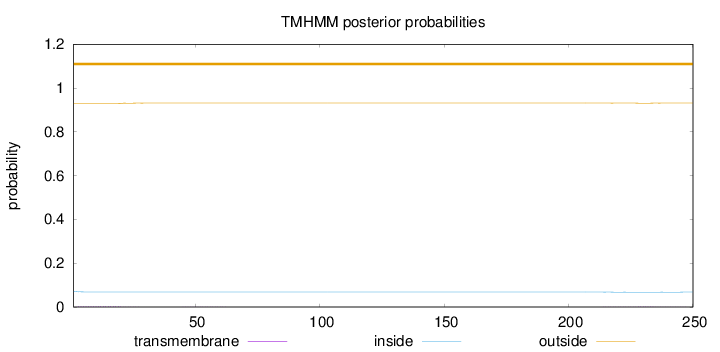
Length:
250
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0901
Exp number, first 60 AAs:
0.05747
Total prob of N-in:
0.07155
outside
1 - 250
Population Genetic Test Statistics
Pi
452.730681
Theta
230.547233
Tajima's D
3.174413
CLR
0.155368
CSRT
0.986300684965752
Interpretation
Uncertain
