Gene
KWMTBOMO16357
Annotation
ribosomal_protein_S11_isoform_1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.268 Mitochondrial Reliability : 1.905 Nuclear Reliability : 1.533
Sequence
CDS
ATGGCGGATCAGACGGAACGCTCATTCCAAAAACAACCTACAGTCTTTCTGAACCGCAAGAAAGGTATTGGTGTGAAGCGGAGCAGAAAACCGTTGAGATACCACAAGGATGTGGGCCTCGGTTTCAAGACTCCCCGTGAGGCGATTGAGGGTACCTACATTGACAAGAAGTGTCCCTTCACTGGCAACGTTTCGATCCGCGGCCGCATCCTCACCGGCGTCGTTCAGAAGATGAAGATGCAGAGAACTATCGTGATCCGCCGCGATTACCTTCACTACCTACCCAAATACAATAGGTTCGAGAAACGGCACAGGAACATGTCCGTACATTTGTCGCCTTGCTTCAGGGACGTGGAGATTGGTGATATTGTAACGATCGGCGAATGCAGACCTTTGTCCAAAACAGTTAGATTCAATGTTTTGAAAGTTTCAAAGGGCAAGGGCTCGAAGAAGTCTTTCAAAAAATTTTAA
Protein
MADQTERSFQKQPTVFLNRKKGIGVKRSRKPLRYHKDVGLGFKTPREAIEGTYIDKKCPFTGNVSIRGRILTGVVQKMKMQRTIVIRRDYLHYLPKYNRFEKRHRNMSVHLSPCFRDVEIGDIVTIGECRPLSKTVRFNVLKVSKGKGSKKSFKKF
Summary
Similarity
Belongs to the universal ribosomal protein uS17 family.
Uniprot
Q5UAN3
D1LYP9
A0A2A4JWD0
E7DZ24
H9JW78
A0A212FIF7
+ More
A0A2H1WZT8 A0A0V0G2Z9 R4G4F1 A0A1W4XHD0 A0A0L7KI72 A0A195BFY4 E2BUQ9 A0A158NBY2 E2A1V8 F4WHW9 A0A2A3EIY7 V9IIV1 A0A088A637 A0A1B6D1G5 A0A194QG42 A0A067R1D4 V5H199 A0A1B6LER8 A0A1B6GPY9 A0A1B6JFT5 A0A0C9Q6X0 Q4GXS7 Q66SW3 A0A0N7Z945 A0A3G1T1N3 E9J9H0 A0A023FAQ2 A0A0L7R1U1 E7DZ23 A0A343EU20 A0A1D1YEK5 A0A195FK25 S4NVN8 G0ZEE2 A2I420 Q5MGJ8 A0A026X3I2 A0A0K2CU27 Q95P67 K7J3W9 C6KI60 I4DM74 I4DIJ8 Q56FI3 A6YPG5 D1LYM5 A0A109NI10 A0A0P5SMI5 A0A2H8TLW2 A0A0M9A8U8 A0A1S4EAV4 D1M804 V5GXT4 C4WS14 A0A0P5N5G5 A0A0P5ULV1 A0A0P5UL75 A0A0P5E3D8 A0A0P5IKT5 A0A0N8ARF6 A0A0P5CRL4 A0A0P5CAS8 A0A0P5Y325 A0A2S2QBJ3 A0A0P5FTC0 A0A0P5AZ60 A0A0P4YLM6 E9GBH6 A0A0N7ZWL7 A0A1B6EAB7 A0A0P5AZ04 A0A0P5AHM3 A0A0P5XS16 A0A171B9V5 A0A0L8I641 A0A0P5W2U3 Q4GXS8 A0A069DPD9 A0A076L0Y8 A2IAD8 A0A1S3IEK6 E0W2J2 A0A1S3CZ68 A0A0A9WDK0 A0A224Z8R4 A0A131YSI9 A0A076KUI1 A0A183IGL6 H2ZXK8 J3JUY0 S4RV01 Q86G63 A0A131XPT7 A0A1W4X744
A0A2H1WZT8 A0A0V0G2Z9 R4G4F1 A0A1W4XHD0 A0A0L7KI72 A0A195BFY4 E2BUQ9 A0A158NBY2 E2A1V8 F4WHW9 A0A2A3EIY7 V9IIV1 A0A088A637 A0A1B6D1G5 A0A194QG42 A0A067R1D4 V5H199 A0A1B6LER8 A0A1B6GPY9 A0A1B6JFT5 A0A0C9Q6X0 Q4GXS7 Q66SW3 A0A0N7Z945 A0A3G1T1N3 E9J9H0 A0A023FAQ2 A0A0L7R1U1 E7DZ23 A0A343EU20 A0A1D1YEK5 A0A195FK25 S4NVN8 G0ZEE2 A2I420 Q5MGJ8 A0A026X3I2 A0A0K2CU27 Q95P67 K7J3W9 C6KI60 I4DM74 I4DIJ8 Q56FI3 A6YPG5 D1LYM5 A0A109NI10 A0A0P5SMI5 A0A2H8TLW2 A0A0M9A8U8 A0A1S4EAV4 D1M804 V5GXT4 C4WS14 A0A0P5N5G5 A0A0P5ULV1 A0A0P5UL75 A0A0P5E3D8 A0A0P5IKT5 A0A0N8ARF6 A0A0P5CRL4 A0A0P5CAS8 A0A0P5Y325 A0A2S2QBJ3 A0A0P5FTC0 A0A0P5AZ60 A0A0P4YLM6 E9GBH6 A0A0N7ZWL7 A0A1B6EAB7 A0A0P5AZ04 A0A0P5AHM3 A0A0P5XS16 A0A171B9V5 A0A0L8I641 A0A0P5W2U3 Q4GXS8 A0A069DPD9 A0A076L0Y8 A2IAD8 A0A1S3IEK6 E0W2J2 A0A1S3CZ68 A0A0A9WDK0 A0A224Z8R4 A0A131YSI9 A0A076KUI1 A0A183IGL6 H2ZXK8 J3JUY0 S4RV01 Q86G63 A0A131XPT7 A0A1W4X744
Pubmed
EMBL
AY769325
AAV34867.1
GU084323
ACY95356.1
NWSH01000450
PCG76325.1
+ More
HQ424727 ADT80671.1 BABH01025469 AGBW02008391 OWR53516.1 ODYU01012349 SOQ58620.1 GECL01003667 JAP02457.1 GAHY01000826 JAA76684.1 JTDY01009683 KOB63048.1 KQ976500 KYM83092.1 GL450743 EFN80564.1 ADTU01011477 GL435806 EFN72597.1 GL888170 EGI66198.1 EGI66199.1 KZ288229 PBC31687.1 JR047988 AEY60552.1 GEDC01017749 JAS19549.1 KQ459054 KPJ03920.1 KK852779 KDR16697.1 GALX01001853 JAB66613.1 GEBQ01017789 JAT22188.1 GECZ01005290 JAS64479.1 GECU01009667 JAS98039.1 GBYB01009853 JAG79620.1 AM048946 CAJ17183.1 AY706955 AY769326 AAU11818.1 AAV34868.1 GDKW01001571 JAI55024.1 MG992456 AXY94894.1 GL769313 EFZ10538.1 GBBI01000593 JAC18119.1 KQ414667 KOC64803.1 HQ424726 ADT80670.1 MF067293 ASM61891.1 GDJX01014853 JAT53083.1 KQ981491 KYN41020.1 GAIX01012877 JAA79683.1 JF265020 AEL28851.1 EF070523 ABM55589.1 AY829788 AAV91402.1 KK107036 EZA61959.1 KT218667 ALA09387.1 AF379640 AAK59928.1 PCG76326.1 AAZX01006133 GQ222274 ACS92720.1 AK402392 BAM19014.1 AK401116 BAM17738.1 AY961517 AAX62419.1 EF638976 ABR27861.1 GU084299 ACY95332.1 KC507308 AHB12431.1 GDIP01141362 JAL62352.1 GFXV01003136 MBW14941.1 KQ435719 KOX78826.1 GU120428 ACY71278.1 GALX01001854 JAB66612.1 ABLF02040572 AK340006 BAH70684.1 GDIQ01146847 JAL04879.1 GDIP01111164 JAL92550.1 GDIP01128813 JAL74901.1 GDIP01165655 JAJ57747.1 GDIQ01236716 JAK15009.1 GDIQ01250211 JAK01514.1 GDIP01167650 JAJ55752.1 GDIP01173798 JAJ49604.1 GDIP01063664 JAM40051.1 GGMS01005359 MBY74562.1 GDIQ01251713 JAK00012.1 GDIP01196356 JAJ27046.1 GDIP01227703 JAI95698.1 GL732538 EFX82959.1 GDIP01205401 JAJ18001.1 GEDC01002412 JAS34886.1 GDIP01192054 GDIP01192053 JAJ31348.1 GDIP01199056 JAJ24346.1 GDIP01069419 JAM34296.1 GEMB01000094 JAS03017.1 KQ416442 KOF96968.1 GDIP01092566 JAM11149.1 AM048945 CAJ17182.1 GBGD01003338 JAC85551.1 KF433792 AII98113.1 EF179452 ABM55458.1 DS235878 EEB19848.1 GBHO01040654 GBRD01001046 GDHC01000783 JAG02950.1 JAG64775.1 JAQ17846.1 GFPF01011804 MAA22950.1 GEDV01006999 JAP81558.1 KF433321 AII97645.1 UZAM01007389 VDO98813.1 AFYH01076246 AFYH01076247 BT127045 AEE62007.1 AY241966 AAO92287.1 GEFH01000209 JAP68372.1
HQ424727 ADT80671.1 BABH01025469 AGBW02008391 OWR53516.1 ODYU01012349 SOQ58620.1 GECL01003667 JAP02457.1 GAHY01000826 JAA76684.1 JTDY01009683 KOB63048.1 KQ976500 KYM83092.1 GL450743 EFN80564.1 ADTU01011477 GL435806 EFN72597.1 GL888170 EGI66198.1 EGI66199.1 KZ288229 PBC31687.1 JR047988 AEY60552.1 GEDC01017749 JAS19549.1 KQ459054 KPJ03920.1 KK852779 KDR16697.1 GALX01001853 JAB66613.1 GEBQ01017789 JAT22188.1 GECZ01005290 JAS64479.1 GECU01009667 JAS98039.1 GBYB01009853 JAG79620.1 AM048946 CAJ17183.1 AY706955 AY769326 AAU11818.1 AAV34868.1 GDKW01001571 JAI55024.1 MG992456 AXY94894.1 GL769313 EFZ10538.1 GBBI01000593 JAC18119.1 KQ414667 KOC64803.1 HQ424726 ADT80670.1 MF067293 ASM61891.1 GDJX01014853 JAT53083.1 KQ981491 KYN41020.1 GAIX01012877 JAA79683.1 JF265020 AEL28851.1 EF070523 ABM55589.1 AY829788 AAV91402.1 KK107036 EZA61959.1 KT218667 ALA09387.1 AF379640 AAK59928.1 PCG76326.1 AAZX01006133 GQ222274 ACS92720.1 AK402392 BAM19014.1 AK401116 BAM17738.1 AY961517 AAX62419.1 EF638976 ABR27861.1 GU084299 ACY95332.1 KC507308 AHB12431.1 GDIP01141362 JAL62352.1 GFXV01003136 MBW14941.1 KQ435719 KOX78826.1 GU120428 ACY71278.1 GALX01001854 JAB66612.1 ABLF02040572 AK340006 BAH70684.1 GDIQ01146847 JAL04879.1 GDIP01111164 JAL92550.1 GDIP01128813 JAL74901.1 GDIP01165655 JAJ57747.1 GDIQ01236716 JAK15009.1 GDIQ01250211 JAK01514.1 GDIP01167650 JAJ55752.1 GDIP01173798 JAJ49604.1 GDIP01063664 JAM40051.1 GGMS01005359 MBY74562.1 GDIQ01251713 JAK00012.1 GDIP01196356 JAJ27046.1 GDIP01227703 JAI95698.1 GL732538 EFX82959.1 GDIP01205401 JAJ18001.1 GEDC01002412 JAS34886.1 GDIP01192054 GDIP01192053 JAJ31348.1 GDIP01199056 JAJ24346.1 GDIP01069419 JAM34296.1 GEMB01000094 JAS03017.1 KQ416442 KOF96968.1 GDIP01092566 JAM11149.1 AM048945 CAJ17182.1 GBGD01003338 JAC85551.1 KF433792 AII98113.1 EF179452 ABM55458.1 DS235878 EEB19848.1 GBHO01040654 GBRD01001046 GDHC01000783 JAG02950.1 JAG64775.1 JAQ17846.1 GFPF01011804 MAA22950.1 GEDV01006999 JAP81558.1 KF433321 AII97645.1 UZAM01007389 VDO98813.1 AFYH01076246 AFYH01076247 BT127045 AEE62007.1 AY241966 AAO92287.1 GEFH01000209 JAP68372.1
Proteomes
UP000218220
UP000005204
UP000007151
UP000192223
UP000037510
UP000078540
+ More
UP000008237 UP000005205 UP000000311 UP000007755 UP000242457 UP000005203 UP000053268 UP000027135 UP000053825 UP000078541 UP000053097 UP000002358 UP000053105 UP000079169 UP000007819 UP000000305 UP000053454 UP000085678 UP000009046 UP000050793 UP000008672 UP000245300
UP000008237 UP000005205 UP000000311 UP000007755 UP000242457 UP000005203 UP000053268 UP000027135 UP000053825 UP000078541 UP000053097 UP000002358 UP000053105 UP000079169 UP000007819 UP000000305 UP000053454 UP000085678 UP000009046 UP000050793 UP000008672 UP000245300
Interpro
SUPFAM
SSF50249
SSF50249
ProteinModelPortal
Q5UAN3
D1LYP9
A0A2A4JWD0
E7DZ24
H9JW78
A0A212FIF7
+ More
A0A2H1WZT8 A0A0V0G2Z9 R4G4F1 A0A1W4XHD0 A0A0L7KI72 A0A195BFY4 E2BUQ9 A0A158NBY2 E2A1V8 F4WHW9 A0A2A3EIY7 V9IIV1 A0A088A637 A0A1B6D1G5 A0A194QG42 A0A067R1D4 V5H199 A0A1B6LER8 A0A1B6GPY9 A0A1B6JFT5 A0A0C9Q6X0 Q4GXS7 Q66SW3 A0A0N7Z945 A0A3G1T1N3 E9J9H0 A0A023FAQ2 A0A0L7R1U1 E7DZ23 A0A343EU20 A0A1D1YEK5 A0A195FK25 S4NVN8 G0ZEE2 A2I420 Q5MGJ8 A0A026X3I2 A0A0K2CU27 Q95P67 K7J3W9 C6KI60 I4DM74 I4DIJ8 Q56FI3 A6YPG5 D1LYM5 A0A109NI10 A0A0P5SMI5 A0A2H8TLW2 A0A0M9A8U8 A0A1S4EAV4 D1M804 V5GXT4 C4WS14 A0A0P5N5G5 A0A0P5ULV1 A0A0P5UL75 A0A0P5E3D8 A0A0P5IKT5 A0A0N8ARF6 A0A0P5CRL4 A0A0P5CAS8 A0A0P5Y325 A0A2S2QBJ3 A0A0P5FTC0 A0A0P5AZ60 A0A0P4YLM6 E9GBH6 A0A0N7ZWL7 A0A1B6EAB7 A0A0P5AZ04 A0A0P5AHM3 A0A0P5XS16 A0A171B9V5 A0A0L8I641 A0A0P5W2U3 Q4GXS8 A0A069DPD9 A0A076L0Y8 A2IAD8 A0A1S3IEK6 E0W2J2 A0A1S3CZ68 A0A0A9WDK0 A0A224Z8R4 A0A131YSI9 A0A076KUI1 A0A183IGL6 H2ZXK8 J3JUY0 S4RV01 Q86G63 A0A131XPT7 A0A1W4X744
A0A2H1WZT8 A0A0V0G2Z9 R4G4F1 A0A1W4XHD0 A0A0L7KI72 A0A195BFY4 E2BUQ9 A0A158NBY2 E2A1V8 F4WHW9 A0A2A3EIY7 V9IIV1 A0A088A637 A0A1B6D1G5 A0A194QG42 A0A067R1D4 V5H199 A0A1B6LER8 A0A1B6GPY9 A0A1B6JFT5 A0A0C9Q6X0 Q4GXS7 Q66SW3 A0A0N7Z945 A0A3G1T1N3 E9J9H0 A0A023FAQ2 A0A0L7R1U1 E7DZ23 A0A343EU20 A0A1D1YEK5 A0A195FK25 S4NVN8 G0ZEE2 A2I420 Q5MGJ8 A0A026X3I2 A0A0K2CU27 Q95P67 K7J3W9 C6KI60 I4DM74 I4DIJ8 Q56FI3 A6YPG5 D1LYM5 A0A109NI10 A0A0P5SMI5 A0A2H8TLW2 A0A0M9A8U8 A0A1S4EAV4 D1M804 V5GXT4 C4WS14 A0A0P5N5G5 A0A0P5ULV1 A0A0P5UL75 A0A0P5E3D8 A0A0P5IKT5 A0A0N8ARF6 A0A0P5CRL4 A0A0P5CAS8 A0A0P5Y325 A0A2S2QBJ3 A0A0P5FTC0 A0A0P5AZ60 A0A0P4YLM6 E9GBH6 A0A0N7ZWL7 A0A1B6EAB7 A0A0P5AZ04 A0A0P5AHM3 A0A0P5XS16 A0A171B9V5 A0A0L8I641 A0A0P5W2U3 Q4GXS8 A0A069DPD9 A0A076L0Y8 A2IAD8 A0A1S3IEK6 E0W2J2 A0A1S3CZ68 A0A0A9WDK0 A0A224Z8R4 A0A131YSI9 A0A076KUI1 A0A183IGL6 H2ZXK8 J3JUY0 S4RV01 Q86G63 A0A131XPT7 A0A1W4X744
PDB
6MTE
E-value=5.63861e-58,
Score=562
Ontologies
KEGG
GO
PANTHER
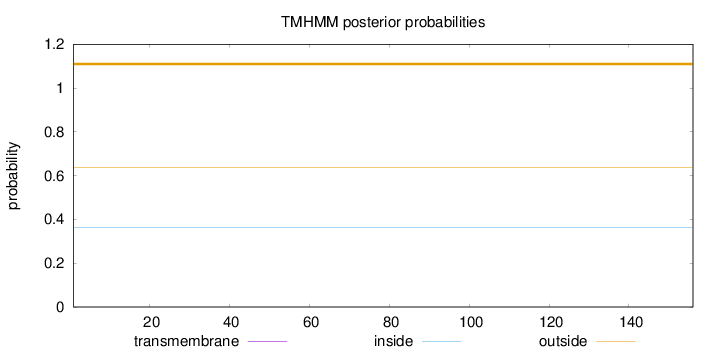
Topology
Length:
156
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000650000000000001
Exp number, first 60 AAs:
4e-05
Total prob of N-in:
0.36296
outside
1 - 156
Population Genetic Test Statistics
Pi
279.294908
Theta
196.903808
Tajima's D
1.33791
CLR
0.000808
CSRT
0.747112644367782
Interpretation
Uncertain
