Gene
KWMTBOMO16348
Pre Gene Modal
BGIBMGA013755
Annotation
PREDICTED:_lachesin-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.124
Sequence
CDS
ATGGCGGAGAATTTTAGGTGCGTGAGAGCAGACGTAGACACTAACGCCCTTGCGCACTTGTCAGTATCCAGGGAGGAAAAGTTAGACTTTCTTCCCTGTACAGCAGGAGGAAAACCGAACTTTCGGTGTACAGCCCACTACGGAGAAGATAGTGTGGGTTCCTTTCCGTACTTCGAGTACAACGTACCGAGGAATGTGACCACAGTAGTCGGACAGACCGCCTTCCTCCACTGCAGGGTAGAGCAACTAGGGGACAAAGCTGTCAGCTGGATCAGGAAGAGGGATCTCCACATACTCACGGCTGGGATTTTGACTTACACATCCGACCAGAGGTTCCAGGTTATTCGGCCTGAGAAGTCAGAAAATTGGACACTGCAGATTAAATTCCCACAGGAGAGAGACGCCGGGATATACGAGTGCCAAGTCAACACGGAACCAAAGATGTCTTTGGCATTTCAACTTAATGTCGTTGAAGCAAAAGCACACATCCTTGGCCCAGCAGATCTGTACGTTAAGACAGGAAGCTCCCTGTCTTTAACTTGTATCCTAAGTCAAGGCCCACACGATCTGGGAACCATATTTTGGTATAAGGGATCAAACCTAATAGAATATAAGGAAGTCGAGGAGAATGAAGTTGCTGAGGAGCCCAGACTTCATCTTAAAACGGAATGGACGGAGCAGCTCACGTCCAGGTTGAGGATAGACAAGCTAATGCCAGGTGACAGCGGAAACTACAGCTGCGTGCCCACTATGGCGGAAGCAGCTTCTGTGAACGTTCACGTCATAAATGGTGAGCACCCGGCAGCAATGCAGCACGGCAACGCCAACACAGCATCACCGTGTGCTCTATCCGTTAATCTTCTCATCTCCCTATACTGCACTTTAGCCACTATATACACGAACACAATCGATGGGCTTAGATGA
Protein
MAENFRCVRADVDTNALAHLSVSREEKLDFLPCTAGGKPNFRCTAHYGEDSVGSFPYFEYNVPRNVTTVVGQTAFLHCRVEQLGDKAVSWIRKRDLHILTAGILTYTSDQRFQVIRPEKSENWTLQIKFPQERDAGIYECQVNTEPKMSLAFQLNVVEAKAHILGPADLYVKTGSSLSLTCILSQGPHDLGTIFWYKGSNLIEYKEVEENEVAEEPRLHLKTEWTEQLTSRLRIDKLMPGDSGNYSCVPTMAEAASVNVHVINGEHPAAMQHGNANTASPCALSVNLLISLYCTLATIYTNTIDGLR
Summary
Uniprot
H9JW42
A0A2W1BIH2
A0A0N1INJ1
A0A2A4JQW8
A0A194PWH4
A0A3S2NDV2
+ More
A0A212EK51 A0A0L7LQB4 A0A1W4WKQ1 A0A1Y1KM36 A0A1W4WLN9 A0A1W4WAL7 D6WQC3 K7J3K8 A0A084W7M1 A0A182W7L9 A0A1S4GW35 A0A182INM9 A0A182PJR9 A0A182X1E9 A0A182M5J8 A0A182IED0 A0A182NTF9 A0A182L2P0 A0A182RJE5 A0A182TN06 W8B7U9 A0A084WJD5 A0A1I8N3M9 A0A182LX90 A0A182WB78 A0A182QCK5 A0A0C9QVT6 A0A182F1B0 A0A182NLY3 A0A1I8P141 B3MIS5 A0A182QZZ0 A0A182HPC2 B4PA21 B3NJX6 A0A1W4VI23 Q8T603 A0A182VP11 A0A0J9RH12 A1ZBY3 A0A1J1J206 B4QF47 B4HQN9 B4KQP6 Q7Q9S5 A0A182WB79 B4ME85 A0A2S2QTF8 Q17KF5 A0A3B0JET8 A0A182LB76 A0A182QG71 A0A0R3NN95 A0A2S2NJI4 A0A182NLY2 B4J970 A0A0Q9WWE4 A0A0Q9W0P6 A0A0M4EVA2 B5E168 Q7Q385 A0A182YJ02 A0A182K2P5 A0A182RUE1 W5J871 Q7Q9S4 A0A182YJ03 A0A182PV21 W5J150 A0A182GPY0 A0A182H4P8 A0A182HDE7 A0A182GLY2 B4HWY0 Q179I1 B4P140 A0A0J9R0G7 B4G6Y0 A0A1W4VT51 A0A336KD98 A0A3L8DGK6
A0A212EK51 A0A0L7LQB4 A0A1W4WKQ1 A0A1Y1KM36 A0A1W4WLN9 A0A1W4WAL7 D6WQC3 K7J3K8 A0A084W7M1 A0A182W7L9 A0A1S4GW35 A0A182INM9 A0A182PJR9 A0A182X1E9 A0A182M5J8 A0A182IED0 A0A182NTF9 A0A182L2P0 A0A182RJE5 A0A182TN06 W8B7U9 A0A084WJD5 A0A1I8N3M9 A0A182LX90 A0A182WB78 A0A182QCK5 A0A0C9QVT6 A0A182F1B0 A0A182NLY3 A0A1I8P141 B3MIS5 A0A182QZZ0 A0A182HPC2 B4PA21 B3NJX6 A0A1W4VI23 Q8T603 A0A182VP11 A0A0J9RH12 A1ZBY3 A0A1J1J206 B4QF47 B4HQN9 B4KQP6 Q7Q9S5 A0A182WB79 B4ME85 A0A2S2QTF8 Q17KF5 A0A3B0JET8 A0A182LB76 A0A182QG71 A0A0R3NN95 A0A2S2NJI4 A0A182NLY2 B4J970 A0A0Q9WWE4 A0A0Q9W0P6 A0A0M4EVA2 B5E168 Q7Q385 A0A182YJ02 A0A182K2P5 A0A182RUE1 W5J871 Q7Q9S4 A0A182YJ03 A0A182PV21 W5J150 A0A182GPY0 A0A182H4P8 A0A182HDE7 A0A182GLY2 B4HWY0 Q179I1 B4P140 A0A0J9R0G7 B4G6Y0 A0A1W4VT51 A0A336KD98 A0A3L8DGK6
Pubmed
19121390
28756777
26354079
22118469
26227816
28004739
+ More
18362917 19820115 20075255 24438588 12364791 20966253 24495485 25315136 17994087 17550304 11978823 30688651 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17510324 15632085 25244985 20920257 23761445 26483478 30249741
18362917 19820115 20075255 24438588 12364791 20966253 24495485 25315136 17994087 17550304 11978823 30688651 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17510324 15632085 25244985 20920257 23761445 26483478 30249741
EMBL
BABH01025407
BABH01025408
KZ150101
PZC73505.1
KQ461053
KPJ09944.1
+ More
NWSH01000852 PCG73843.1 KQ459588 KPI97726.1 RSAL01000180 RVE44935.1 AGBW02014312 OWR41892.1 JTDY01000339 KOB77632.1 GEZM01081402 JAV61648.1 KQ971354 EFA06091.2 ATLV01021267 KE525315 KFB46215.1 AAAB01008964 AXCM01002382 APCN01005201 GAMC01011878 JAB94677.1 ATLV01024017 KE525348 KFB50329.1 AXCM01018170 AXCN02000706 GBYB01007859 JAG77626.1 CH902619 EDV35985.2 AXCN02000954 APCN01002967 CM000158 EDW91352.1 CH954179 EDV55351.1 AF489698 AAM18045.1 CM002911 KMY95323.1 AE013599 BT050529 AAS64909.2 ACJ13236.1 AHN56440.1 CVRI01000063 CRL04889.1 CM000362 EDX07958.1 CH480816 EDW48742.1 CH933808 EDW08215.2 AAAB01008900 EAA09508.4 CH940662 EDW58850.1 GGMS01011730 MBY80933.1 CH477224 EAT47196.1 OUUW01000001 SPP73810.1 CM000071 KRT02370.1 GGMR01004473 MBY17092.1 CH916367 EDW02445.1 CH964282 KRG00316.1 KRF78557.1 CP012524 ALC41520.1 EDY69084.2 EAA12592.4 ADMH02002104 ETN59065.1 EAA09317.4 ADMH02002200 ETN57867.1 JXUM01079533 JXUM01079534 JXUM01079535 KQ563132 KXJ74429.1 JXUM01109976 JXUM01109977 KQ565370 KXJ71043.1 JXUM01034276 JXUM01034277 JXUM01034278 KQ561018 KXJ80015.1 JXUM01073826 JXUM01073827 JXUM01073828 KQ562797 KXJ75108.1 CH480818 EDW52525.1 CH477349 EAT42909.1 CM000157 EDW88015.2 CM002910 KMY89646.1 KMY89648.1 CH479180 EDW28299.1 UFQS01000083 UFQT01000083 SSW99060.1 SSX19442.1 QOIP01000008 RLU19580.1
NWSH01000852 PCG73843.1 KQ459588 KPI97726.1 RSAL01000180 RVE44935.1 AGBW02014312 OWR41892.1 JTDY01000339 KOB77632.1 GEZM01081402 JAV61648.1 KQ971354 EFA06091.2 ATLV01021267 KE525315 KFB46215.1 AAAB01008964 AXCM01002382 APCN01005201 GAMC01011878 JAB94677.1 ATLV01024017 KE525348 KFB50329.1 AXCM01018170 AXCN02000706 GBYB01007859 JAG77626.1 CH902619 EDV35985.2 AXCN02000954 APCN01002967 CM000158 EDW91352.1 CH954179 EDV55351.1 AF489698 AAM18045.1 CM002911 KMY95323.1 AE013599 BT050529 AAS64909.2 ACJ13236.1 AHN56440.1 CVRI01000063 CRL04889.1 CM000362 EDX07958.1 CH480816 EDW48742.1 CH933808 EDW08215.2 AAAB01008900 EAA09508.4 CH940662 EDW58850.1 GGMS01011730 MBY80933.1 CH477224 EAT47196.1 OUUW01000001 SPP73810.1 CM000071 KRT02370.1 GGMR01004473 MBY17092.1 CH916367 EDW02445.1 CH964282 KRG00316.1 KRF78557.1 CP012524 ALC41520.1 EDY69084.2 EAA12592.4 ADMH02002104 ETN59065.1 EAA09317.4 ADMH02002200 ETN57867.1 JXUM01079533 JXUM01079534 JXUM01079535 KQ563132 KXJ74429.1 JXUM01109976 JXUM01109977 KQ565370 KXJ71043.1 JXUM01034276 JXUM01034277 JXUM01034278 KQ561018 KXJ80015.1 JXUM01073826 JXUM01073827 JXUM01073828 KQ562797 KXJ75108.1 CH480818 EDW52525.1 CH477349 EAT42909.1 CM000157 EDW88015.2 CM002910 KMY89646.1 KMY89648.1 CH479180 EDW28299.1 UFQS01000083 UFQT01000083 SSW99060.1 SSX19442.1 QOIP01000008 RLU19580.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000283053
UP000007151
+ More
UP000037510 UP000192223 UP000007266 UP000002358 UP000030765 UP000075920 UP000075880 UP000075885 UP000076407 UP000075883 UP000075840 UP000075884 UP000075882 UP000075900 UP000075902 UP000095301 UP000075886 UP000069272 UP000095300 UP000007801 UP000002282 UP000008711 UP000192221 UP000075903 UP000000803 UP000183832 UP000000304 UP000001292 UP000009192 UP000007062 UP000008792 UP000008820 UP000268350 UP000001819 UP000001070 UP000007798 UP000092553 UP000076408 UP000075881 UP000000673 UP000069940 UP000249989 UP000008744 UP000279307
UP000037510 UP000192223 UP000007266 UP000002358 UP000030765 UP000075920 UP000075880 UP000075885 UP000076407 UP000075883 UP000075840 UP000075884 UP000075882 UP000075900 UP000075902 UP000095301 UP000075886 UP000069272 UP000095300 UP000007801 UP000002282 UP000008711 UP000192221 UP000075903 UP000000803 UP000183832 UP000000304 UP000001292 UP000009192 UP000007062 UP000008792 UP000008820 UP000268350 UP000001819 UP000001070 UP000007798 UP000092553 UP000076408 UP000075881 UP000000673 UP000069940 UP000249989 UP000008744 UP000279307
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
H9JW42
A0A2W1BIH2
A0A0N1INJ1
A0A2A4JQW8
A0A194PWH4
A0A3S2NDV2
+ More
A0A212EK51 A0A0L7LQB4 A0A1W4WKQ1 A0A1Y1KM36 A0A1W4WLN9 A0A1W4WAL7 D6WQC3 K7J3K8 A0A084W7M1 A0A182W7L9 A0A1S4GW35 A0A182INM9 A0A182PJR9 A0A182X1E9 A0A182M5J8 A0A182IED0 A0A182NTF9 A0A182L2P0 A0A182RJE5 A0A182TN06 W8B7U9 A0A084WJD5 A0A1I8N3M9 A0A182LX90 A0A182WB78 A0A182QCK5 A0A0C9QVT6 A0A182F1B0 A0A182NLY3 A0A1I8P141 B3MIS5 A0A182QZZ0 A0A182HPC2 B4PA21 B3NJX6 A0A1W4VI23 Q8T603 A0A182VP11 A0A0J9RH12 A1ZBY3 A0A1J1J206 B4QF47 B4HQN9 B4KQP6 Q7Q9S5 A0A182WB79 B4ME85 A0A2S2QTF8 Q17KF5 A0A3B0JET8 A0A182LB76 A0A182QG71 A0A0R3NN95 A0A2S2NJI4 A0A182NLY2 B4J970 A0A0Q9WWE4 A0A0Q9W0P6 A0A0M4EVA2 B5E168 Q7Q385 A0A182YJ02 A0A182K2P5 A0A182RUE1 W5J871 Q7Q9S4 A0A182YJ03 A0A182PV21 W5J150 A0A182GPY0 A0A182H4P8 A0A182HDE7 A0A182GLY2 B4HWY0 Q179I1 B4P140 A0A0J9R0G7 B4G6Y0 A0A1W4VT51 A0A336KD98 A0A3L8DGK6
A0A212EK51 A0A0L7LQB4 A0A1W4WKQ1 A0A1Y1KM36 A0A1W4WLN9 A0A1W4WAL7 D6WQC3 K7J3K8 A0A084W7M1 A0A182W7L9 A0A1S4GW35 A0A182INM9 A0A182PJR9 A0A182X1E9 A0A182M5J8 A0A182IED0 A0A182NTF9 A0A182L2P0 A0A182RJE5 A0A182TN06 W8B7U9 A0A084WJD5 A0A1I8N3M9 A0A182LX90 A0A182WB78 A0A182QCK5 A0A0C9QVT6 A0A182F1B0 A0A182NLY3 A0A1I8P141 B3MIS5 A0A182QZZ0 A0A182HPC2 B4PA21 B3NJX6 A0A1W4VI23 Q8T603 A0A182VP11 A0A0J9RH12 A1ZBY3 A0A1J1J206 B4QF47 B4HQN9 B4KQP6 Q7Q9S5 A0A182WB79 B4ME85 A0A2S2QTF8 Q17KF5 A0A3B0JET8 A0A182LB76 A0A182QG71 A0A0R3NN95 A0A2S2NJI4 A0A182NLY2 B4J970 A0A0Q9WWE4 A0A0Q9W0P6 A0A0M4EVA2 B5E168 Q7Q385 A0A182YJ02 A0A182K2P5 A0A182RUE1 W5J871 Q7Q9S4 A0A182YJ03 A0A182PV21 W5J150 A0A182GPY0 A0A182H4P8 A0A182HDE7 A0A182GLY2 B4HWY0 Q179I1 B4P140 A0A0J9R0G7 B4G6Y0 A0A1W4VT51 A0A336KD98 A0A3L8DGK6
PDB
6EG1
E-value=1.71519e-63,
Score=614
Ontologies
GO
PANTHER
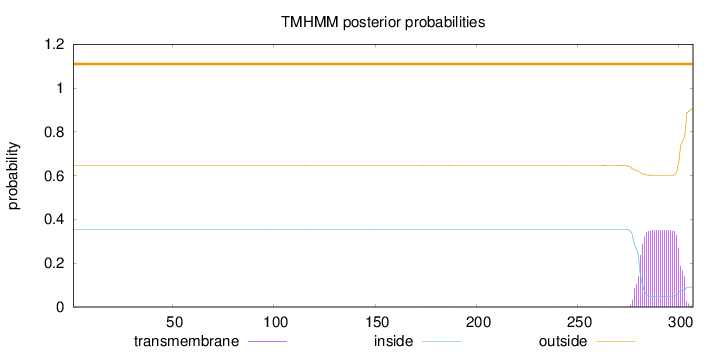
Topology
Length:
307
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.63441
Exp number, first 60 AAs:
1e-05
Total prob of N-in:
0.35437
outside
1 - 307
Population Genetic Test Statistics
Pi
210.110703
Theta
183.278133
Tajima's D
0.798524
CLR
0.267677
CSRT
0.597170141492925
Interpretation
Uncertain
