Gene
KWMTBOMO16346 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013756
Annotation
chitin_deacetylase_17?_partial_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.668 PlasmaMembrane Reliability : 2.096
Sequence
CDS
ATGAAGAGGCTGTCGGTTTTGTGCTCTTTGCTCCTCGTGGCTGCGGCTCTTGGCACTGAGCTGCCATTGGCTACACCATGTGACGAAGAGGCTTGTAAGCTCCCGGACTGCAGATGCTCGTCCACTAATATTCCTGGAGGATTACGAGCCAGAGACACTCCACAGTTTGTAACCGTAACGTTCGATGACGGTATCAATGTCATTAACATTGAGACATACCGTGAGGTCCTTTACGGCCGCAGCAACTCGAACCGCTGCCCAGCCGGAGCTACCTTCTACGTCAGCCACGAGTACACAAACTATCAGCTGGTCAACGAACTCTACAATAGAGGCTTCGAGATTGCTCTACATTCGATCAGTCACAGAACCCCCCAGGCGTTCTGGGCTGATGCCACGTATCAAAACTTGGTCCAGGAAATTGGAGATCAGAAAAGACAAATGGCGCATTTTGCTAGCATCCCCGCTAGTGCAATTAAAGGTGTGCGTATCCCCTTCCTTCAGATGTCCGGTAACACAAGTTTCCAGGTTATGGCAGATTTTGACTTACTGTACGACTGCACCTGGCCCACAACTGCCTTGACTAATCCAGGTCTATGGCCTTACACTCTTCACCACGAATCTATTCAGGACTGCATTATTCCTCCATGCCCAACTGCGTCAATTCCTGGACCGTGGGTGCTCCCTATGATCAGCTGGAGAGACCTCAACAATTTCCCTTGCTCCATGGTGGATGGTTGCTTCTTCACGCCAGACCGTACCGACGAGGAAGGATGGTTCAAGTTTATCCTGACCAACTTCGAGAGACACTACCTCGGGAACAGAGCCCCGTTCGGATTCTTCGTCCACGAATGGTTCATTTCGTCAAACCCCGCCATCAAACGCGCTTTTGTCAGATTCATGGATATCATTAACAACTTAAATGATGTCTTCATGGTGAACTCAGCTGAAGTCATCGATTGGGTGAAGAACCCCGTCCCGATCGATCGATACCGTCAGCAGCAGTGCAAATTTACTATGCCATCAATTTGTAGACCATCGTTCTGCGGGCCACTAACGGGAACTCATAATCAGCTGTCTTACTACATGACGATTTGCAACACGTGTCCCAGGAATTACCCCTGGGTAGGAAATCCGCTTGGACAGTAA
Protein
MKRLSVLCSLLLVAAALGTELPLATPCDEEACKLPDCRCSSTNIPGGLRARDTPQFVTVTFDDGINVINIETYREVLYGRSNSNRCPAGATFYVSHEYTNYQLVNELYNRGFEIALHSISHRTPQAFWADATYQNLVQEIGDQKRQMAHFASIPASAIKGVRIPFLQMSGNTSFQVMADFDLLYDCTWPTTALTNPGLWPYTLHHESIQDCIIPPCPTASIPGPWVLPMISWRDLNNFPCSMVDGCFFTPDRTDEEGWFKFILTNFERHYLGNRAPFGFFVHEWFISSNPAIKRAFVRFMDIINNLNDVFMVNSAEVIDWVKNPVPIDRYRQQQCKFTMPSICRPSFCGPLTGTHNQLSYYMTICNTCPRNYPWVGNPLGQ
Summary
Uniprot
H9JW43
A0A0F6T2X6
A0A2A4JBX6
D2WPC5
A0A2W1BJR7
B1NLD6
+ More
A0A2A4JB10 D2WPC6 B6CME5 A0A2W1BFD3 A0A0N1PFV6 A0A194PXI1 A0A194PWM8 F6K727 A0A3S5FTY2 B2ZGH5 A0A1E1WFV4 A0A2H1WQD6 F8SU34 H9JW44 Q3B9L9 A0A212F1F8 A0A0L7K4B0 A0A3Q7ZTN7 A0A0L7K492 H9JW45 L7X1K8 B6CME6 A0A1E1W3P0 A0A2H1WDN2 B0WIN7 A0A1Q3FLZ8 A8W495 A0A1B0G7T6 A0A1A9ZI54 A0A1A9UGX9 A0A336M0N0 A0A0L0BRI3 A0A0A1XAF6 A0A1B0B4G1 A0A0T6B3I1 A0A1A9XWF8 A0A1I8N882 A0A0M4ETT8 B4KQF6 B4LKH6 B4P8G5 A0A1I8PRQ5 W8C7K2 B4J554 A0A0K8W047 B4HMI5 B4QAQ8 A0A1A9WWS3 Q4V447 A1ZAQ7 A0A3B0J1A7 A0A2Z2Z010 A0A1W4VCQ6 A0A034WG48 B3NNU3 B3MD53 Q28XD6 B5DUL3 B4N5S4 B4GIS8 T1P9N1 A0A0P5BKX5 A0A0P4ZTP9 A0A0P5ZVM8 A0A0N8EIL8 A0A0P6BU74 A0A0P5Y6M4 A0A0C9QAD3 A0A2B4SFR0 A0A0P5IWN6 A0A0P6FFR2 A0A0P5ZY23 A0A0P5AXC9 A0A1D2MDX5 E9GTI2 A0A1B0CLD7 A0A0P5Y8S2 A0A226EA36 A0A182J0W8 A0A0P6GID6 A0A087UZJ7 A7T0W4 A0A0N8EFX0 A0A0P4XP64 A0A1D2NCB7 A0A3R7MXU9 A0A0A1X2J9 A0A182H803 A0A0P6CYF9 A0A0P6ACL1 A0A182UVJ7
A0A2A4JB10 D2WPC6 B6CME5 A0A2W1BFD3 A0A0N1PFV6 A0A194PXI1 A0A194PWM8 F6K727 A0A3S5FTY2 B2ZGH5 A0A1E1WFV4 A0A2H1WQD6 F8SU34 H9JW44 Q3B9L9 A0A212F1F8 A0A0L7K4B0 A0A3Q7ZTN7 A0A0L7K492 H9JW45 L7X1K8 B6CME6 A0A1E1W3P0 A0A2H1WDN2 B0WIN7 A0A1Q3FLZ8 A8W495 A0A1B0G7T6 A0A1A9ZI54 A0A1A9UGX9 A0A336M0N0 A0A0L0BRI3 A0A0A1XAF6 A0A1B0B4G1 A0A0T6B3I1 A0A1A9XWF8 A0A1I8N882 A0A0M4ETT8 B4KQF6 B4LKH6 B4P8G5 A0A1I8PRQ5 W8C7K2 B4J554 A0A0K8W047 B4HMI5 B4QAQ8 A0A1A9WWS3 Q4V447 A1ZAQ7 A0A3B0J1A7 A0A2Z2Z010 A0A1W4VCQ6 A0A034WG48 B3NNU3 B3MD53 Q28XD6 B5DUL3 B4N5S4 B4GIS8 T1P9N1 A0A0P5BKX5 A0A0P4ZTP9 A0A0P5ZVM8 A0A0N8EIL8 A0A0P6BU74 A0A0P5Y6M4 A0A0C9QAD3 A0A2B4SFR0 A0A0P5IWN6 A0A0P6FFR2 A0A0P5ZY23 A0A0P5AXC9 A0A1D2MDX5 E9GTI2 A0A1B0CLD7 A0A0P5Y8S2 A0A226EA36 A0A182J0W8 A0A0P6GID6 A0A087UZJ7 A7T0W4 A0A0N8EFX0 A0A0P4XP64 A0A1D2NCB7 A0A3R7MXU9 A0A0A1X2J9 A0A182H803 A0A0P6CYF9 A0A0P6ACL1 A0A182UVJ7
Pubmed
19121390
20032185
28756777
18314941
18760362
26354079
+ More
16203204 22118469 26227816 23674305 18362917 19820115 26108605 25830018 25315136 17994087 17550304 24495485 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 15632085 27289101 21292972 17615350 26483478
16203204 22118469 26227816 23674305 18362917 19820115 26108605 25830018 25315136 17994087 17550304 24495485 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 15632085 27289101 21292972 17615350 26483478
EMBL
BABH01025382
KP744648
AKD49099.1
NWSH01002064
PCG69266.1
GQ411190
+ More
ADB43611.1 KZ150137 PZC72970.1 EF600051 ABU98616.1 PCG69265.1 GQ411191 ADB43612.1 EU325545 ACB54935.1 PZC72971.1 KQ461053 KPJ09940.1 KQ459588 KPI97728.1 KPI97727.1 HM357864 AEA76330.1 KR363154 AMY98414.1 EU660852 ACD37362.1 GDQN01005164 JAT85890.1 ODYU01010231 SOQ55207.1 HQ680621 AEI30869.1 AY966402 AAY46199.1 AGBW02010886 OWR47572.1 JTDY01011406 KOB56570.1 KP159288 AKO73677.1 KOB56571.1 BABH01025381 KC469895 AGC92658.1 EU325546 ACB54936.1 GDQN01009456 JAT81598.1 ODYU01007948 SOQ51161.1 DS231951 EDS28614.1 GFDL01006480 JAV28565.1 EU190492 KQ971330 ABW74152.1 EFA00992.1 CCAG010016895 UFQT01000391 SSX23886.1 JRES01001465 KNC22685.1 GBXI01006013 JAD08279.1 JXJN01008300 LJIG01016008 KRT81890.1 CP012524 ALC40987.1 CH933808 EDW10296.1 CH940648 EDW60697.1 CM000158 EDW92182.1 GAMC01003574 JAC02982.1 CH916367 EDW00680.1 GDHF01007807 JAI44507.1 CH480816 EDW48253.1 CM000362 CM002911 EDX07459.1 KMY94474.1 BT023159 AAY55575.1 AE013599 BT088821 AAF57892.1 ACS35599.1 AHN56315.1 OUUW01000001 SPP74934.1 KY681052 AUP42579.1 GAKP01005288 JAC53664.1 CH954179 EDV55650.1 CH902619 EDV37386.1 CM000071 EAL26380.1 CH673921 EDY71579.1 CH964154 EDW79713.1 CH479183 EDW35413.1 KA645437 AFP60066.1 GDIP01183140 JAJ40262.1 GDIP01208687 JAJ14715.1 GDIP01201838 GDIP01037909 LRGB01000027 JAM65806.1 KZS21268.1 GDIQ01023124 JAN71613.1 GDIP01010132 JAM93583.1 GDIP01063514 JAM40201.1 GBYB01011398 JAG81165.1 LSMT01000104 PFX27397.1 GDIQ01208818 JAK42907.1 GDIQ01060520 JAN34217.1 GDIP01037059 JAM66656.1 GDIP01193044 JAJ30358.1 LJIJ01001635 ODM91190.1 GL732564 EFX77211.1 AJWK01017126 GDIP01062160 JAM41555.1 LNIX01000005 OXA54405.1 GDIQ01032802 JAN61935.1 KK122473 KFM82786.1 DS470055 EDO30402.1 GDIQ01030708 JAN64029.1 GDIP01239768 JAI83633.1 LJIJ01000093 ODN02879.1 QCYY01002184 ROT72283.1 GBXI01008718 JAD05574.1 JXUM01028413 JXUM01028414 JXUM01028415 KQ560821 KXJ80757.1 GDIQ01085739 JAN08998.1 GDIP01036241 JAM67474.1
ADB43611.1 KZ150137 PZC72970.1 EF600051 ABU98616.1 PCG69265.1 GQ411191 ADB43612.1 EU325545 ACB54935.1 PZC72971.1 KQ461053 KPJ09940.1 KQ459588 KPI97728.1 KPI97727.1 HM357864 AEA76330.1 KR363154 AMY98414.1 EU660852 ACD37362.1 GDQN01005164 JAT85890.1 ODYU01010231 SOQ55207.1 HQ680621 AEI30869.1 AY966402 AAY46199.1 AGBW02010886 OWR47572.1 JTDY01011406 KOB56570.1 KP159288 AKO73677.1 KOB56571.1 BABH01025381 KC469895 AGC92658.1 EU325546 ACB54936.1 GDQN01009456 JAT81598.1 ODYU01007948 SOQ51161.1 DS231951 EDS28614.1 GFDL01006480 JAV28565.1 EU190492 KQ971330 ABW74152.1 EFA00992.1 CCAG010016895 UFQT01000391 SSX23886.1 JRES01001465 KNC22685.1 GBXI01006013 JAD08279.1 JXJN01008300 LJIG01016008 KRT81890.1 CP012524 ALC40987.1 CH933808 EDW10296.1 CH940648 EDW60697.1 CM000158 EDW92182.1 GAMC01003574 JAC02982.1 CH916367 EDW00680.1 GDHF01007807 JAI44507.1 CH480816 EDW48253.1 CM000362 CM002911 EDX07459.1 KMY94474.1 BT023159 AAY55575.1 AE013599 BT088821 AAF57892.1 ACS35599.1 AHN56315.1 OUUW01000001 SPP74934.1 KY681052 AUP42579.1 GAKP01005288 JAC53664.1 CH954179 EDV55650.1 CH902619 EDV37386.1 CM000071 EAL26380.1 CH673921 EDY71579.1 CH964154 EDW79713.1 CH479183 EDW35413.1 KA645437 AFP60066.1 GDIP01183140 JAJ40262.1 GDIP01208687 JAJ14715.1 GDIP01201838 GDIP01037909 LRGB01000027 JAM65806.1 KZS21268.1 GDIQ01023124 JAN71613.1 GDIP01010132 JAM93583.1 GDIP01063514 JAM40201.1 GBYB01011398 JAG81165.1 LSMT01000104 PFX27397.1 GDIQ01208818 JAK42907.1 GDIQ01060520 JAN34217.1 GDIP01037059 JAM66656.1 GDIP01193044 JAJ30358.1 LJIJ01001635 ODM91190.1 GL732564 EFX77211.1 AJWK01017126 GDIP01062160 JAM41555.1 LNIX01000005 OXA54405.1 GDIQ01032802 JAN61935.1 KK122473 KFM82786.1 DS470055 EDO30402.1 GDIQ01030708 JAN64029.1 GDIP01239768 JAI83633.1 LJIJ01000093 ODN02879.1 QCYY01002184 ROT72283.1 GBXI01008718 JAD05574.1 JXUM01028413 JXUM01028414 JXUM01028415 KQ560821 KXJ80757.1 GDIQ01085739 JAN08998.1 GDIP01036241 JAM67474.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000002320 UP000007266 UP000092444 UP000092445 UP000078200 UP000037069 UP000092460 UP000092443 UP000095301 UP000092553 UP000009192 UP000008792 UP000002282 UP000095300 UP000001070 UP000001292 UP000000304 UP000091820 UP000000803 UP000268350 UP000192221 UP000008711 UP000007801 UP000001819 UP000007798 UP000008744 UP000076858 UP000225706 UP000094527 UP000000305 UP000092461 UP000198287 UP000075880 UP000054359 UP000001593 UP000283509 UP000069940 UP000249989 UP000075903
UP000002320 UP000007266 UP000092444 UP000092445 UP000078200 UP000037069 UP000092460 UP000092443 UP000095301 UP000092553 UP000009192 UP000008792 UP000002282 UP000095300 UP000001070 UP000001292 UP000000304 UP000091820 UP000000803 UP000268350 UP000192221 UP000008711 UP000007801 UP000001819 UP000007798 UP000008744 UP000076858 UP000225706 UP000094527 UP000000305 UP000092461 UP000198287 UP000075880 UP000054359 UP000001593 UP000283509 UP000069940 UP000249989 UP000075903
Interpro
ProteinModelPortal
H9JW43
A0A0F6T2X6
A0A2A4JBX6
D2WPC5
A0A2W1BJR7
B1NLD6
+ More
A0A2A4JB10 D2WPC6 B6CME5 A0A2W1BFD3 A0A0N1PFV6 A0A194PXI1 A0A194PWM8 F6K727 A0A3S5FTY2 B2ZGH5 A0A1E1WFV4 A0A2H1WQD6 F8SU34 H9JW44 Q3B9L9 A0A212F1F8 A0A0L7K4B0 A0A3Q7ZTN7 A0A0L7K492 H9JW45 L7X1K8 B6CME6 A0A1E1W3P0 A0A2H1WDN2 B0WIN7 A0A1Q3FLZ8 A8W495 A0A1B0G7T6 A0A1A9ZI54 A0A1A9UGX9 A0A336M0N0 A0A0L0BRI3 A0A0A1XAF6 A0A1B0B4G1 A0A0T6B3I1 A0A1A9XWF8 A0A1I8N882 A0A0M4ETT8 B4KQF6 B4LKH6 B4P8G5 A0A1I8PRQ5 W8C7K2 B4J554 A0A0K8W047 B4HMI5 B4QAQ8 A0A1A9WWS3 Q4V447 A1ZAQ7 A0A3B0J1A7 A0A2Z2Z010 A0A1W4VCQ6 A0A034WG48 B3NNU3 B3MD53 Q28XD6 B5DUL3 B4N5S4 B4GIS8 T1P9N1 A0A0P5BKX5 A0A0P4ZTP9 A0A0P5ZVM8 A0A0N8EIL8 A0A0P6BU74 A0A0P5Y6M4 A0A0C9QAD3 A0A2B4SFR0 A0A0P5IWN6 A0A0P6FFR2 A0A0P5ZY23 A0A0P5AXC9 A0A1D2MDX5 E9GTI2 A0A1B0CLD7 A0A0P5Y8S2 A0A226EA36 A0A182J0W8 A0A0P6GID6 A0A087UZJ7 A7T0W4 A0A0N8EFX0 A0A0P4XP64 A0A1D2NCB7 A0A3R7MXU9 A0A0A1X2J9 A0A182H803 A0A0P6CYF9 A0A0P6ACL1 A0A182UVJ7
A0A2A4JB10 D2WPC6 B6CME5 A0A2W1BFD3 A0A0N1PFV6 A0A194PXI1 A0A194PWM8 F6K727 A0A3S5FTY2 B2ZGH5 A0A1E1WFV4 A0A2H1WQD6 F8SU34 H9JW44 Q3B9L9 A0A212F1F8 A0A0L7K4B0 A0A3Q7ZTN7 A0A0L7K492 H9JW45 L7X1K8 B6CME6 A0A1E1W3P0 A0A2H1WDN2 B0WIN7 A0A1Q3FLZ8 A8W495 A0A1B0G7T6 A0A1A9ZI54 A0A1A9UGX9 A0A336M0N0 A0A0L0BRI3 A0A0A1XAF6 A0A1B0B4G1 A0A0T6B3I1 A0A1A9XWF8 A0A1I8N882 A0A0M4ETT8 B4KQF6 B4LKH6 B4P8G5 A0A1I8PRQ5 W8C7K2 B4J554 A0A0K8W047 B4HMI5 B4QAQ8 A0A1A9WWS3 Q4V447 A1ZAQ7 A0A3B0J1A7 A0A2Z2Z010 A0A1W4VCQ6 A0A034WG48 B3NNU3 B3MD53 Q28XD6 B5DUL3 B4N5S4 B4GIS8 T1P9N1 A0A0P5BKX5 A0A0P4ZTP9 A0A0P5ZVM8 A0A0N8EIL8 A0A0P6BU74 A0A0P5Y6M4 A0A0C9QAD3 A0A2B4SFR0 A0A0P5IWN6 A0A0P6FFR2 A0A0P5ZY23 A0A0P5AXC9 A0A1D2MDX5 E9GTI2 A0A1B0CLD7 A0A0P5Y8S2 A0A226EA36 A0A182J0W8 A0A0P6GID6 A0A087UZJ7 A7T0W4 A0A0N8EFX0 A0A0P4XP64 A0A1D2NCB7 A0A3R7MXU9 A0A0A1X2J9 A0A182H803 A0A0P6CYF9 A0A0P6ACL1 A0A182UVJ7
PDB
5Z34
E-value=0,
Score=1974
Ontologies
GO
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.999271
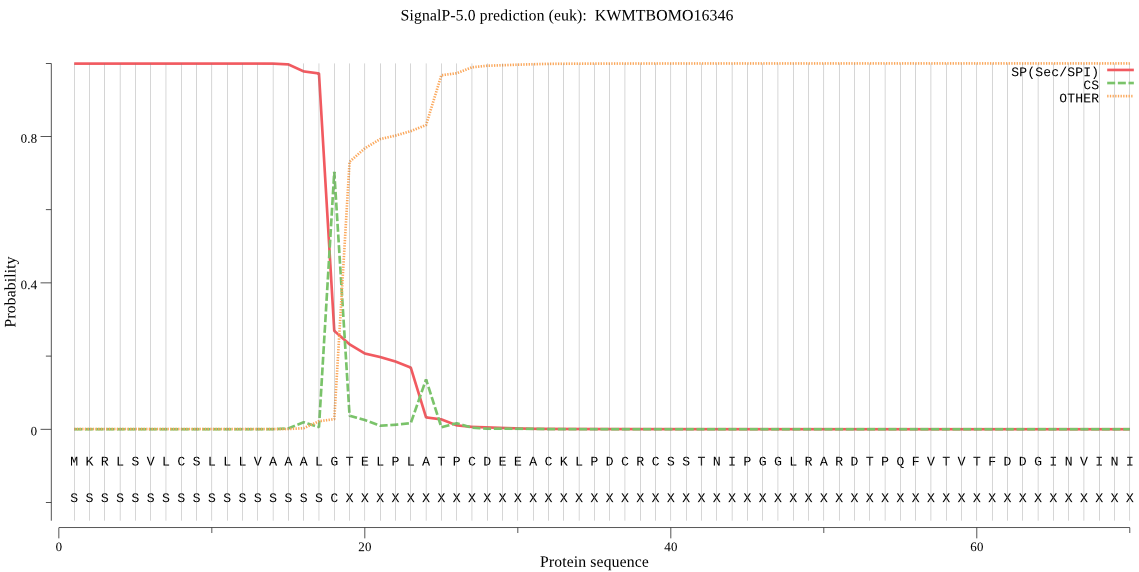
Length:
381
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.22961
Exp number, first 60 AAs:
1.20438
Total prob of N-in:
0.06668
outside
1 - 381

Population Genetic Test Statistics
Pi
275.391763
Theta
194.235432
Tajima's D
1.436881
CLR
0.449939
CSRT
0.769811509424529
Interpretation
Uncertain
