Gene
KWMTBOMO16334
Pre Gene Modal
BGIBMGA013783
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_protein_Wnt-5b_[Bombyx_mori]
Full name
Protein Wnt
Location in the cell
Extracellular Reliability : 3.147
Sequence
CDS
ATGAACAGACACAACAACGAGGCTGGTAGACGGGCGGTGATAGCAAAATCACGGGTGACTTGCAAATGTCACGGAGTGTCGGGATCCTGCAGCCTGATCACGTGTTGGCAGCAGCTAGCGTCTTTTAGGGAGATCGGCGATTATCTGAGAGACCGATACGAAGGCGCAACAGAAGTGAAAGTTTCGAGACGAGGGAAGCTGCGTCTCAGAAATCCGAGGTACAGCCTGCCGACAGCACAAGATCTGGTGTACTTGGAAGAGTCGCCGAATTACTGTGTCAAGAATGAGTCTTTGGGCTCCCTCGGTACTTCTGGCCGAGAATGCAACAGGACTTCCTTAGGCTTAGACGGGTGCTCATTACTGTGCTGCGGAAGAGGTTACAACACTAAAAGAATAATGATGAAAGAACGATGTCAGTGCAAGTTCCACTGGTGCTGCCACGTGAAATGCAATACTTGTGTCAAATCCACCGAACTTTATACTTGTAAATAA
Protein
MNRHNNEAGRRAVIAKSRVTCKCHGVSGSCSLITCWQQLASFREIGDYLRDRYEGATEVKVSRRGKLRLRNPRYSLPTAQDLVYLEESPNYCVKNESLGSLGTSGRECNRTSLGLDGCSLLCCGRGYNTKRIMMKERCQCKFHWCCHVKCNTCVKSTELYTCK
Summary
Description
Ligand for members of the frizzled family of seven transmembrane receptors.
Similarity
Belongs to the Wnt family.
Feature
chain Protein Wnt
Uniprot
M4B151
A0A2W1BP63
A0A2A4JRJ8
A0A0N0P9T2
A0A0N0PDY9
A0A212F380
+ More
A0A3S2LE46 A0A2H1VID8 A0A0M8ZWT5 A0A0L7R138 A0A2A3EHV6 A0A088ANQ7 A0A154NY47 A0A232ENR7 K7IRG4 A0A158NE00 F4X1T4 A0A195D7Z0 E2AX67 A0A0S1NEZ4 A0A067QPQ6 A0A0A9XWT0 A0A195C4V9 E2BFD9 X5JAY7 A0A1B6DN32 A0A1B6D4P7 A0A1Y1L4G4 E9HP53 E0VAI0 T1HF57 A0A1B6K9R9 A0A0P6BBJ1 A0A0K8TFX1 D6WRU0 A0A2S2NW04 A0A2R7WYN5 A0A026W0U9 A0A151X4W7 A0A2H8TF98 Q75PH7 N6TD29 J9JT78 A0A195BPR1 U4URI0 A0A195ERL6 Q8T395 T1J4F4 A0A0P4WA74 A0A1L2A1P3 A0A0P6IBR6 A0A182T8L7 A0A097ZRN9 A0A1I8NSH3 A0A182VSR4 A0A182NNM7 A0A182RAG1 A0A1B0EVX1 A0A182K4H1 A0A182GBU5 A0A182L7V1 A0A087U247 W8B6E3 A0A182MSA1 T1GWS8 B4I6T6 B7PYK7 A0A2M4CIM5 A0A2M4CIQ3 Q1DH67 A0A0L0CHV7 A0A1I8M7F0 B3MWR5 A0A034V1K5 A0A084W5N0 A0A132AGW6 A0A182YN19 A0A182JEQ7 A0A1S4G2M1 A0A182QI97 A0A182PTW3 B4HBK1 Q7Q0K5 A0A0A1X731 A0A0A1X250 A0A3Q0IIE6 B4LPF4 B4J6G3 A0A3P3ZKP2 B4KLT8 A0A0Q9XIJ7 A0A1S4H3J8 A0A3Q0IIA3 A0A0R1EDS7 B4MQU9 B0X7A8 B4Q2Z5 A0A0Q5THZ3 A0A182I5E9 A0A0K8V7G0
A0A3S2LE46 A0A2H1VID8 A0A0M8ZWT5 A0A0L7R138 A0A2A3EHV6 A0A088ANQ7 A0A154NY47 A0A232ENR7 K7IRG4 A0A158NE00 F4X1T4 A0A195D7Z0 E2AX67 A0A0S1NEZ4 A0A067QPQ6 A0A0A9XWT0 A0A195C4V9 E2BFD9 X5JAY7 A0A1B6DN32 A0A1B6D4P7 A0A1Y1L4G4 E9HP53 E0VAI0 T1HF57 A0A1B6K9R9 A0A0P6BBJ1 A0A0K8TFX1 D6WRU0 A0A2S2NW04 A0A2R7WYN5 A0A026W0U9 A0A151X4W7 A0A2H8TF98 Q75PH7 N6TD29 J9JT78 A0A195BPR1 U4URI0 A0A195ERL6 Q8T395 T1J4F4 A0A0P4WA74 A0A1L2A1P3 A0A0P6IBR6 A0A182T8L7 A0A097ZRN9 A0A1I8NSH3 A0A182VSR4 A0A182NNM7 A0A182RAG1 A0A1B0EVX1 A0A182K4H1 A0A182GBU5 A0A182L7V1 A0A087U247 W8B6E3 A0A182MSA1 T1GWS8 B4I6T6 B7PYK7 A0A2M4CIM5 A0A2M4CIQ3 Q1DH67 A0A0L0CHV7 A0A1I8M7F0 B3MWR5 A0A034V1K5 A0A084W5N0 A0A132AGW6 A0A182YN19 A0A182JEQ7 A0A1S4G2M1 A0A182QI97 A0A182PTW3 B4HBK1 Q7Q0K5 A0A0A1X731 A0A0A1X250 A0A3Q0IIE6 B4LPF4 B4J6G3 A0A3P3ZKP2 B4KLT8 A0A0Q9XIJ7 A0A1S4H3J8 A0A3Q0IIA3 A0A0R1EDS7 B4MQU9 B0X7A8 B4Q2Z5 A0A0Q5THZ3 A0A182I5E9 A0A0K8V7G0
Pubmed
19121390
28756777
26354079
22118469
28648823
20075255
+ More
21347285 21719571 20798317 24845553 25401762 24398121 28004739 21292972 20566863 26823975 18362917 19820115 24508170 30249741 26561354 23537049 11874919 29325710 29567137 26483478 20966253 24495485 17994087 17510324 26108605 25315136 25348373 24438588 26555130 25244985 12364791 25830018 18057021 17550304
21347285 21719571 20798317 24845553 25401762 24398121 28004739 21292972 20566863 26823975 18362917 19820115 24508170 30249741 26561354 23537049 11874919 29325710 29567137 26483478 20966253 24495485 17994087 17510324 26108605 25315136 25348373 24438588 26555130 25244985 12364791 25830018 18057021 17550304
EMBL
BABH01025365
BABH01025366
BABH01025367
KZ149970
PZC76071.1
NWSH01000767
+ More
PCG74328.1 KQ459166 KPJ03558.1 KQ459992 KPJ18597.1 AGBW02010607 OWR48186.1 RSAL01000029 RVE51762.1 ODYU01002715 SOQ40571.1 KQ435833 KOX71666.1 KQ414668 KOC64559.1 KZ288254 PBC30779.1 KQ434782 KZC04543.1 NNAY01003079 OXU20013.1 ADTU01013080 ADTU01013081 GL888551 EGI59591.1 KQ981153 KYN09005.1 GL443520 EFN61950.1 KT818598 ALL53297.1 KK853153 KDR10544.1 GBHO01019235 JAG24369.1 KQ978251 KYM95907.1 GL448009 EFN85599.1 HG529220 CDI40111.1 GEDC01010215 JAS27083.1 GEDC01016619 JAS20679.1 GEZM01065069 JAV68573.1 GL732703 EFX66479.1 DS235006 EEB10386.1 ACPB03015483 GEBQ01031785 GEBQ01004158 JAT08192.1 JAT35819.1 GDIP01017713 JAM86002.1 GBRD01001520 GDHC01004452 JAG64301.1 JAQ14177.1 KQ971351 EFA07476.1 GGMR01008623 MBY21242.1 KK856014 PTY24637.1 KK107503 QOIP01000006 EZA49633.1 RLU21728.1 KQ982548 KYQ55310.1 GFXV01000944 MBW12749.1 AB167810 IAAA01025754 BAD12588.1 LAA08095.1 APGK01035285 APGK01035286 APGK01035287 APGK01035288 APGK01035289 KB740923 ENN78244.1 ABLF02040228 KQ976424 KYM88507.1 KB632333 ERL92675.1 KQ981993 KYN30821.1 AJ315946 CAC87041.1 JH431845 GDRN01105948 GDRN01105944 JAI57650.1 KU169897 MF958962 MF416111 ALO81628.1 AWW24763.1 AXN72349.1 GDIQ01007790 JAN86947.1 HG529210 CDI40101.1 AJWK01022156 JXUM01053338 JXUM01062195 KQ562188 KQ561772 KXJ76462.1 KXJ77588.1 KK117793 KFM71436.1 GAMC01017679 JAB88876.1 AXCM01002077 CAQQ02107965 CAQQ02107966 CH480823 EDW56034.1 ABJB010771883 DS820456 EEC11679.1 GGFL01001006 MBW65184.1 GGFL01001005 MBW65183.1 CH899796 EAT32499.1 JRES01000379 KNC31797.1 CH902626 EDV41419.2 GAKP01023306 JAC35650.1 ATLV01020675 ATLV01020676 KE525304 KFB45524.1 JXLN01013914 KPM09690.1 AXCN02001876 CH479257 EDW38779.1 AAAB01008980 EAA14587.4 GBXI01007819 JAD06473.1 GBXI01009095 JAD05197.1 CH940648 EDW61213.2 CH916367 EDW01964.1 LR025119 VAY10397.1 CH933808 EDW09748.1 KRG04882.1 CM000162 KRK06942.1 CH963849 EDW74488.1 DS232445 EDS41861.1 EDX02729.2 CH954180 KQS30059.1 APCN01006407 GDHF01017452 JAI34862.1
PCG74328.1 KQ459166 KPJ03558.1 KQ459992 KPJ18597.1 AGBW02010607 OWR48186.1 RSAL01000029 RVE51762.1 ODYU01002715 SOQ40571.1 KQ435833 KOX71666.1 KQ414668 KOC64559.1 KZ288254 PBC30779.1 KQ434782 KZC04543.1 NNAY01003079 OXU20013.1 ADTU01013080 ADTU01013081 GL888551 EGI59591.1 KQ981153 KYN09005.1 GL443520 EFN61950.1 KT818598 ALL53297.1 KK853153 KDR10544.1 GBHO01019235 JAG24369.1 KQ978251 KYM95907.1 GL448009 EFN85599.1 HG529220 CDI40111.1 GEDC01010215 JAS27083.1 GEDC01016619 JAS20679.1 GEZM01065069 JAV68573.1 GL732703 EFX66479.1 DS235006 EEB10386.1 ACPB03015483 GEBQ01031785 GEBQ01004158 JAT08192.1 JAT35819.1 GDIP01017713 JAM86002.1 GBRD01001520 GDHC01004452 JAG64301.1 JAQ14177.1 KQ971351 EFA07476.1 GGMR01008623 MBY21242.1 KK856014 PTY24637.1 KK107503 QOIP01000006 EZA49633.1 RLU21728.1 KQ982548 KYQ55310.1 GFXV01000944 MBW12749.1 AB167810 IAAA01025754 BAD12588.1 LAA08095.1 APGK01035285 APGK01035286 APGK01035287 APGK01035288 APGK01035289 KB740923 ENN78244.1 ABLF02040228 KQ976424 KYM88507.1 KB632333 ERL92675.1 KQ981993 KYN30821.1 AJ315946 CAC87041.1 JH431845 GDRN01105948 GDRN01105944 JAI57650.1 KU169897 MF958962 MF416111 ALO81628.1 AWW24763.1 AXN72349.1 GDIQ01007790 JAN86947.1 HG529210 CDI40101.1 AJWK01022156 JXUM01053338 JXUM01062195 KQ562188 KQ561772 KXJ76462.1 KXJ77588.1 KK117793 KFM71436.1 GAMC01017679 JAB88876.1 AXCM01002077 CAQQ02107965 CAQQ02107966 CH480823 EDW56034.1 ABJB010771883 DS820456 EEC11679.1 GGFL01001006 MBW65184.1 GGFL01001005 MBW65183.1 CH899796 EAT32499.1 JRES01000379 KNC31797.1 CH902626 EDV41419.2 GAKP01023306 JAC35650.1 ATLV01020675 ATLV01020676 KE525304 KFB45524.1 JXLN01013914 KPM09690.1 AXCN02001876 CH479257 EDW38779.1 AAAB01008980 EAA14587.4 GBXI01007819 JAD06473.1 GBXI01009095 JAD05197.1 CH940648 EDW61213.2 CH916367 EDW01964.1 LR025119 VAY10397.1 CH933808 EDW09748.1 KRG04882.1 CM000162 KRK06942.1 CH963849 EDW74488.1 DS232445 EDS41861.1 EDX02729.2 CH954180 KQS30059.1 APCN01006407 GDHF01017452 JAI34862.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000283053
+ More
UP000053105 UP000053825 UP000242457 UP000005203 UP000076502 UP000215335 UP000002358 UP000005205 UP000007755 UP000078492 UP000000311 UP000027135 UP000078542 UP000008237 UP000000305 UP000009046 UP000015103 UP000007266 UP000053097 UP000279307 UP000075809 UP000019118 UP000007819 UP000078540 UP000030742 UP000078541 UP000075901 UP000095300 UP000075920 UP000075884 UP000075900 UP000092461 UP000075881 UP000069940 UP000249989 UP000075882 UP000054359 UP000075883 UP000015102 UP000001292 UP000001555 UP000008820 UP000037069 UP000095301 UP000007801 UP000030765 UP000076408 UP000075880 UP000075886 UP000075885 UP000008744 UP000007062 UP000079169 UP000008792 UP000001070 UP000009192 UP000002282 UP000007798 UP000002320 UP000008711 UP000075840
UP000053105 UP000053825 UP000242457 UP000005203 UP000076502 UP000215335 UP000002358 UP000005205 UP000007755 UP000078492 UP000000311 UP000027135 UP000078542 UP000008237 UP000000305 UP000009046 UP000015103 UP000007266 UP000053097 UP000279307 UP000075809 UP000019118 UP000007819 UP000078540 UP000030742 UP000078541 UP000075901 UP000095300 UP000075920 UP000075884 UP000075900 UP000092461 UP000075881 UP000069940 UP000249989 UP000075882 UP000054359 UP000075883 UP000015102 UP000001292 UP000001555 UP000008820 UP000037069 UP000095301 UP000007801 UP000030765 UP000076408 UP000075880 UP000075886 UP000075885 UP000008744 UP000007062 UP000079169 UP000008792 UP000001070 UP000009192 UP000002282 UP000007798 UP000002320 UP000008711 UP000075840
PRIDE
ProteinModelPortal
M4B151
A0A2W1BP63
A0A2A4JRJ8
A0A0N0P9T2
A0A0N0PDY9
A0A212F380
+ More
A0A3S2LE46 A0A2H1VID8 A0A0M8ZWT5 A0A0L7R138 A0A2A3EHV6 A0A088ANQ7 A0A154NY47 A0A232ENR7 K7IRG4 A0A158NE00 F4X1T4 A0A195D7Z0 E2AX67 A0A0S1NEZ4 A0A067QPQ6 A0A0A9XWT0 A0A195C4V9 E2BFD9 X5JAY7 A0A1B6DN32 A0A1B6D4P7 A0A1Y1L4G4 E9HP53 E0VAI0 T1HF57 A0A1B6K9R9 A0A0P6BBJ1 A0A0K8TFX1 D6WRU0 A0A2S2NW04 A0A2R7WYN5 A0A026W0U9 A0A151X4W7 A0A2H8TF98 Q75PH7 N6TD29 J9JT78 A0A195BPR1 U4URI0 A0A195ERL6 Q8T395 T1J4F4 A0A0P4WA74 A0A1L2A1P3 A0A0P6IBR6 A0A182T8L7 A0A097ZRN9 A0A1I8NSH3 A0A182VSR4 A0A182NNM7 A0A182RAG1 A0A1B0EVX1 A0A182K4H1 A0A182GBU5 A0A182L7V1 A0A087U247 W8B6E3 A0A182MSA1 T1GWS8 B4I6T6 B7PYK7 A0A2M4CIM5 A0A2M4CIQ3 Q1DH67 A0A0L0CHV7 A0A1I8M7F0 B3MWR5 A0A034V1K5 A0A084W5N0 A0A132AGW6 A0A182YN19 A0A182JEQ7 A0A1S4G2M1 A0A182QI97 A0A182PTW3 B4HBK1 Q7Q0K5 A0A0A1X731 A0A0A1X250 A0A3Q0IIE6 B4LPF4 B4J6G3 A0A3P3ZKP2 B4KLT8 A0A0Q9XIJ7 A0A1S4H3J8 A0A3Q0IIA3 A0A0R1EDS7 B4MQU9 B0X7A8 B4Q2Z5 A0A0Q5THZ3 A0A182I5E9 A0A0K8V7G0
A0A3S2LE46 A0A2H1VID8 A0A0M8ZWT5 A0A0L7R138 A0A2A3EHV6 A0A088ANQ7 A0A154NY47 A0A232ENR7 K7IRG4 A0A158NE00 F4X1T4 A0A195D7Z0 E2AX67 A0A0S1NEZ4 A0A067QPQ6 A0A0A9XWT0 A0A195C4V9 E2BFD9 X5JAY7 A0A1B6DN32 A0A1B6D4P7 A0A1Y1L4G4 E9HP53 E0VAI0 T1HF57 A0A1B6K9R9 A0A0P6BBJ1 A0A0K8TFX1 D6WRU0 A0A2S2NW04 A0A2R7WYN5 A0A026W0U9 A0A151X4W7 A0A2H8TF98 Q75PH7 N6TD29 J9JT78 A0A195BPR1 U4URI0 A0A195ERL6 Q8T395 T1J4F4 A0A0P4WA74 A0A1L2A1P3 A0A0P6IBR6 A0A182T8L7 A0A097ZRN9 A0A1I8NSH3 A0A182VSR4 A0A182NNM7 A0A182RAG1 A0A1B0EVX1 A0A182K4H1 A0A182GBU5 A0A182L7V1 A0A087U247 W8B6E3 A0A182MSA1 T1GWS8 B4I6T6 B7PYK7 A0A2M4CIM5 A0A2M4CIQ3 Q1DH67 A0A0L0CHV7 A0A1I8M7F0 B3MWR5 A0A034V1K5 A0A084W5N0 A0A132AGW6 A0A182YN19 A0A182JEQ7 A0A1S4G2M1 A0A182QI97 A0A182PTW3 B4HBK1 Q7Q0K5 A0A0A1X731 A0A0A1X250 A0A3Q0IIE6 B4LPF4 B4J6G3 A0A3P3ZKP2 B4KLT8 A0A0Q9XIJ7 A0A1S4H3J8 A0A3Q0IIA3 A0A0R1EDS7 B4MQU9 B0X7A8 B4Q2Z5 A0A0Q5THZ3 A0A182I5E9 A0A0K8V7G0
PDB
6AHY
E-value=4.24603e-35,
Score=365
Ontologies
KEGG
PATHWAY
GO
PANTHER
Topology
Subcellular location
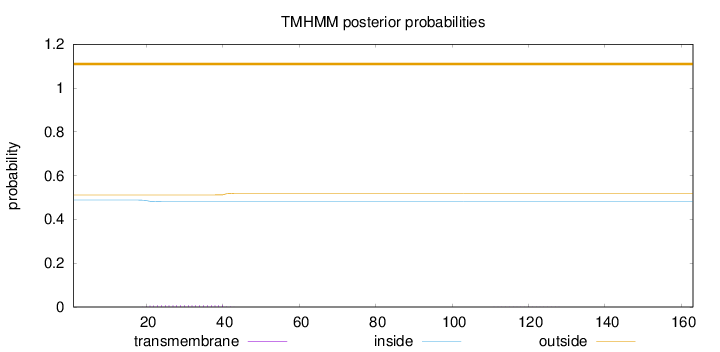
Length:
163
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.162
Exp number, first 60 AAs:
0.15794
Total prob of N-in:
0.48830
outside
1 - 163
Population Genetic Test Statistics
Pi
242.670573
Theta
170.687129
Tajima's D
1.59008
CLR
0.97858
CSRT
0.807459627018649
Interpretation
Uncertain
