Pre Gene Modal
BGIBMGA013777
Annotation
receptor_expression_enhancing_protein_isoform_1_[Bombyx_mori]
Full name
Receptor expression-enhancing protein
Location in the cell
PlasmaMembrane Reliability : 3.939
Sequence
CDS
ATGGCATCCAAATTACAAGAGTATAAAGACAACATAGAACAAAGTCTGAACGACAAAAGCAAGCCATGGACGAAATATTTTGAACTAGCCGAACAAAAGGTCGGCGTGAACAGATTGTACATTTTCTTAGGCTTGGTCGCGTTCACCGGTTTATATTTAGTGTTCGGTTTCGGAGCTGAATTAATATGCAACTCGATCGGCTTCGTGTACCCCGCGTACATGTCTATGAAGGCTCTGGAGTCACCGCAGAAGGATGACGATACAAAATGGCTTACATATTGGGTGGTGTACGCCTGTTTTTCTATCGTGGAGTACTTTTCCGATTTCATCGTCGGCTGGTTTCCATTGTACTGGCTGTTAAAGTGCATCTTCGTGATCTGGTGCTACCTGCCGACTGAATACAACGGGTCCCTCGTGATCTACTACCGTATCATTCGTCCTTACTACCAGAAGCATCATGGTCGTATCGATGATATGGCCAACACAGCTTCCCGTCTGGTGGCAGATACCATCAGAAAGACCAATTAA
Protein
MASKLQEYKDNIEQSLNDKSKPWTKYFELAEQKVGVNRLYIFLGLVAFTGLYLVFGFGAELICNSIGFVYPAYMSMKALESPQKDDDTKWLTYWVVYACFSIVEYFSDFIVGWFPLYWLLKCIFVIWCYLPTEYNGSLVIYYRIIRPYYQKHHGRIDDMANTASRLVADTIRKTN
Summary
Similarity
Belongs to the DP1 family.
Uniprot
Q2F5K7
Q2F5K6
A0A125R9I4
A0A125R9I5
H9JW64
A0A3S2LQJ7
+ More
A0A1E1WM44 A0A2W1BI48 A0A2A4K806 A0A0N1PJ53 S4NXT8 A0A0N1IDF3 A0A212F313 A0A1E1WSZ2 I4DQD7 J3JUL2 D6WQ76 N6UI22 A0A1Y1LGS0 A0A1W4WLW4 A0A1W4WBT0 A0A1B6C5U4 A0A0L7LCF7 A0A067RUB6 A0A0T6AZM5 K7IX69 A0A232F9F3 E2C7V6 A0A0J7LBB9 A0A2J7QS09 E1ZY09 A0A026WT89 A0A195FEM0 V5GF95 E0VH70 U5EW28 A0A0K8UWI9 A0A0C9R406 A0A0A1X874 A0A1S3DQX1 A0A0P4VZ98 R4G485 A0A087ZTL5 A0A2A3E6M7 V9IK43 A0A195AWQ8 A0A158NHT7 A0A0K8TRV3 W8ALM3 A0A195EGB3 A0A0P6JZ62 T1DIV1 A0A336LPK7 A0A224XLU4 A0A1B0CI81 A0A1L8EHN0 A0A151WR26 A0A023F9A2 A0A069DPV7 A0A0M5J380 A0A1B6LC06 A0A1D2NIA8 A0A0V0G3N1 A0A0L0CM13 A0A023EID9 T1DF73 A0A1I8MPB5 Q16VD7 A0A0A9ZF23 A0A195CBL1 A0A1L8DV19 A0A3R7PTC9 A0A1Q3EU70 A0A182LI22 Q7PMR4 A0A182YPH5 B4HR19 B4QFD6 F0JAH0 A1Z9M2 Q8T9H5 A0A0L7R0T1 A0A154NYK2 B3MCZ1 A0A0M9A7M6 A0A0P4WZE4 A0A2M4ACF6 Q16VD6 B4P6N0 A0A1W4VDF7 A0A1B6FIY9 A0A2M3Z4D0 B4LLE8 E9HCE1 A0A1Q3EUI2 A0A0P5YI07 A0A0P5Z508 B3NRC2 D3TN14
A0A1E1WM44 A0A2W1BI48 A0A2A4K806 A0A0N1PJ53 S4NXT8 A0A0N1IDF3 A0A212F313 A0A1E1WSZ2 I4DQD7 J3JUL2 D6WQ76 N6UI22 A0A1Y1LGS0 A0A1W4WLW4 A0A1W4WBT0 A0A1B6C5U4 A0A0L7LCF7 A0A067RUB6 A0A0T6AZM5 K7IX69 A0A232F9F3 E2C7V6 A0A0J7LBB9 A0A2J7QS09 E1ZY09 A0A026WT89 A0A195FEM0 V5GF95 E0VH70 U5EW28 A0A0K8UWI9 A0A0C9R406 A0A0A1X874 A0A1S3DQX1 A0A0P4VZ98 R4G485 A0A087ZTL5 A0A2A3E6M7 V9IK43 A0A195AWQ8 A0A158NHT7 A0A0K8TRV3 W8ALM3 A0A195EGB3 A0A0P6JZ62 T1DIV1 A0A336LPK7 A0A224XLU4 A0A1B0CI81 A0A1L8EHN0 A0A151WR26 A0A023F9A2 A0A069DPV7 A0A0M5J380 A0A1B6LC06 A0A1D2NIA8 A0A0V0G3N1 A0A0L0CM13 A0A023EID9 T1DF73 A0A1I8MPB5 Q16VD7 A0A0A9ZF23 A0A195CBL1 A0A1L8DV19 A0A3R7PTC9 A0A1Q3EU70 A0A182LI22 Q7PMR4 A0A182YPH5 B4HR19 B4QFD6 F0JAH0 A1Z9M2 Q8T9H5 A0A0L7R0T1 A0A154NYK2 B3MCZ1 A0A0M9A7M6 A0A0P4WZE4 A0A2M4ACF6 Q16VD6 B4P6N0 A0A1W4VDF7 A0A1B6FIY9 A0A2M3Z4D0 B4LLE8 E9HCE1 A0A1Q3EUI2 A0A0P5YI07 A0A0P5Z508 B3NRC2 D3TN14
Pubmed
26656276
19121390
28756777
26354079
23622113
22118469
+ More
22651552 22516182 23537049 18362917 19820115 28004739 26227816 24845553 20075255 28648823 20798317 24508170 30249741 20566863 25830018 27129103 21347285 26369729 24495485 24330624 25474469 26334808 27289101 26108605 24945155 25315136 17510324 25401762 26823975 20966253 12364791 14747013 17210077 25244985 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 21292972 20353571
22651552 22516182 23537049 18362917 19820115 28004739 26227816 24845553 20075255 28648823 20798317 24508170 30249741 20566863 25830018 27129103 21347285 26369729 24495485 24330624 25474469 26334808 27289101 26108605 24945155 25315136 17510324 25401762 26823975 20966253 12364791 14747013 17210077 25244985 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 21292972 20353571
EMBL
DQ311416
ABD36360.1
DQ311417
ABD36361.1
KR260842
AMD16373.1
+ More
KR260843 AMD16374.1 BABH01025340 BABH01025341 RSAL01000029 RVE51756.1 GDQN01007024 GDQN01003036 JAT84030.1 JAT88018.1 KZ150109 PZC73375.1 NWSH01000040 PCG80361.1 KQ459992 KPJ18582.1 GAIX01012027 JAA80533.1 KQ459166 KPJ03574.1 AGBW02010634 OWR48138.1 GDQN01004184 GDQN01000956 JAT86870.1 JAT90098.1 AK404059 BAM20127.1 BT126927 KB632331 AEE61889.1 ERL92630.1 KQ971354 EFA06115.1 APGK01019273 APGK01019274 KB740098 ENN81395.1 GEZM01057952 GEZM01057951 JAV72071.1 GEDC01028451 JAS08847.1 JTDY01001713 KOB73099.1 KK852461 KDR23434.1 LJIG01022471 KRT80404.1 AAZX01009916 NNAY01000643 OXU27232.1 GL453466 EFN75991.1 LBMM01000040 KMR05207.1 NEVH01011881 PNF31369.1 GL435164 EFN73928.1 KK107109 QOIP01000008 EZA59158.1 RLU19651.1 KQ981636 KYN38848.1 GALX01005822 JAB62644.1 DS235161 EEB12726.1 GANO01003146 JAB56725.1 GDHF01021426 JAI30888.1 GBYB01011034 JAG80801.1 GBXI01006773 JAD07519.1 GDKW01001734 JAI54861.1 GAHY01001580 JAA75930.1 KZ288368 PBC26819.1 JR048433 AEY60664.1 KQ976725 KYM76606.1 ADTU01016037 GDAI01000725 JAI16878.1 GAMC01019733 JAB86822.1 KQ978957 KYN27278.1 GDIQ01002352 JAN92385.1 GALA01000736 JAA94116.1 UFQS01000102 UFQT01000102 SSW99626.1 SSX20006.1 GFTR01002979 JAW13447.1 AJWK01013083 GFDG01000673 JAV18126.1 KQ982813 KYQ50263.1 GBBI01001219 JAC17493.1 GBGD01003168 JAC85721.1 CP012524 ALC42711.1 GEBQ01018744 GEBQ01011998 JAT21233.1 JAT27979.1 LJIJ01000031 ODN05000.1 GECL01003434 JAP02690.1 JRES01000204 KNC33311.1 GAPW01004907 JAC08691.1 GALA01000737 JAA94115.1 CH477597 EAT38496.1 EJY57811.1 GBHO01001073 GDHC01009884 JAG42531.1 JAQ08745.1 KQ978023 KYM98085.1 GFDF01003949 JAV10135.1 QCYY01001649 ROT76517.1 GFDL01016186 JAV18859.1 AAAB01008968 EAA13329.4 CH480816 EDW47816.1 CM000362 CM002911 EDX07046.1 KMY93695.1 BT126012 ADY17711.1 AE013599 AAF58285.1 AY069293 AAL39438.1 KQ414669 KOC64457.1 KQ434782 KZC04661.1 CH902619 EDV36306.1 KQ435732 KOX77529.1 GDRN01043394 JAI66985.1 GGFK01005128 MBW38449.1 EAT38494.1 CM000158 EDW90982.1 GECZ01019604 JAS50165.1 GGFM01002630 MBW23381.1 CH940648 EDW61900.1 GL732619 EFX70606.1 GFDL01016187 JAV18858.1 GDIP01057419 JAM46296.1 GDIQ01096265 GDIP01050122 LRGB01013148 JAL55461.1 JAM53593.1 KZR99617.1 CH954179 EDV56074.2 EZ422816 ADD19092.1
KR260843 AMD16374.1 BABH01025340 BABH01025341 RSAL01000029 RVE51756.1 GDQN01007024 GDQN01003036 JAT84030.1 JAT88018.1 KZ150109 PZC73375.1 NWSH01000040 PCG80361.1 KQ459992 KPJ18582.1 GAIX01012027 JAA80533.1 KQ459166 KPJ03574.1 AGBW02010634 OWR48138.1 GDQN01004184 GDQN01000956 JAT86870.1 JAT90098.1 AK404059 BAM20127.1 BT126927 KB632331 AEE61889.1 ERL92630.1 KQ971354 EFA06115.1 APGK01019273 APGK01019274 KB740098 ENN81395.1 GEZM01057952 GEZM01057951 JAV72071.1 GEDC01028451 JAS08847.1 JTDY01001713 KOB73099.1 KK852461 KDR23434.1 LJIG01022471 KRT80404.1 AAZX01009916 NNAY01000643 OXU27232.1 GL453466 EFN75991.1 LBMM01000040 KMR05207.1 NEVH01011881 PNF31369.1 GL435164 EFN73928.1 KK107109 QOIP01000008 EZA59158.1 RLU19651.1 KQ981636 KYN38848.1 GALX01005822 JAB62644.1 DS235161 EEB12726.1 GANO01003146 JAB56725.1 GDHF01021426 JAI30888.1 GBYB01011034 JAG80801.1 GBXI01006773 JAD07519.1 GDKW01001734 JAI54861.1 GAHY01001580 JAA75930.1 KZ288368 PBC26819.1 JR048433 AEY60664.1 KQ976725 KYM76606.1 ADTU01016037 GDAI01000725 JAI16878.1 GAMC01019733 JAB86822.1 KQ978957 KYN27278.1 GDIQ01002352 JAN92385.1 GALA01000736 JAA94116.1 UFQS01000102 UFQT01000102 SSW99626.1 SSX20006.1 GFTR01002979 JAW13447.1 AJWK01013083 GFDG01000673 JAV18126.1 KQ982813 KYQ50263.1 GBBI01001219 JAC17493.1 GBGD01003168 JAC85721.1 CP012524 ALC42711.1 GEBQ01018744 GEBQ01011998 JAT21233.1 JAT27979.1 LJIJ01000031 ODN05000.1 GECL01003434 JAP02690.1 JRES01000204 KNC33311.1 GAPW01004907 JAC08691.1 GALA01000737 JAA94115.1 CH477597 EAT38496.1 EJY57811.1 GBHO01001073 GDHC01009884 JAG42531.1 JAQ08745.1 KQ978023 KYM98085.1 GFDF01003949 JAV10135.1 QCYY01001649 ROT76517.1 GFDL01016186 JAV18859.1 AAAB01008968 EAA13329.4 CH480816 EDW47816.1 CM000362 CM002911 EDX07046.1 KMY93695.1 BT126012 ADY17711.1 AE013599 AAF58285.1 AY069293 AAL39438.1 KQ414669 KOC64457.1 KQ434782 KZC04661.1 CH902619 EDV36306.1 KQ435732 KOX77529.1 GDRN01043394 JAI66985.1 GGFK01005128 MBW38449.1 EAT38494.1 CM000158 EDW90982.1 GECZ01019604 JAS50165.1 GGFM01002630 MBW23381.1 CH940648 EDW61900.1 GL732619 EFX70606.1 GFDL01016187 JAV18858.1 GDIP01057419 JAM46296.1 GDIQ01096265 GDIP01050122 LRGB01013148 JAL55461.1 JAM53593.1 KZR99617.1 CH954179 EDV56074.2 EZ422816 ADD19092.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000030742 UP000007266 UP000019118 UP000192223 UP000037510 UP000027135 UP000002358 UP000215335 UP000008237 UP000036403 UP000235965 UP000000311 UP000053097 UP000279307 UP000078541 UP000009046 UP000079169 UP000005203 UP000242457 UP000078540 UP000005205 UP000078492 UP000092461 UP000075809 UP000092553 UP000094527 UP000037069 UP000095301 UP000008820 UP000078542 UP000283509 UP000075882 UP000007062 UP000076408 UP000001292 UP000000304 UP000000803 UP000053825 UP000076502 UP000007801 UP000053105 UP000002282 UP000192221 UP000008792 UP000000305 UP000076858 UP000008711
UP000030742 UP000007266 UP000019118 UP000192223 UP000037510 UP000027135 UP000002358 UP000215335 UP000008237 UP000036403 UP000235965 UP000000311 UP000053097 UP000279307 UP000078541 UP000009046 UP000079169 UP000005203 UP000242457 UP000078540 UP000005205 UP000078492 UP000092461 UP000075809 UP000092553 UP000094527 UP000037069 UP000095301 UP000008820 UP000078542 UP000283509 UP000075882 UP000007062 UP000076408 UP000001292 UP000000304 UP000000803 UP000053825 UP000076502 UP000007801 UP000053105 UP000002282 UP000192221 UP000008792 UP000000305 UP000076858 UP000008711
Pfam
PF03134 TB2_DP1_HVA22
Interpro
IPR004345
TB2_DP1_HVA22
ProteinModelPortal
Q2F5K7
Q2F5K6
A0A125R9I4
A0A125R9I5
H9JW64
A0A3S2LQJ7
+ More
A0A1E1WM44 A0A2W1BI48 A0A2A4K806 A0A0N1PJ53 S4NXT8 A0A0N1IDF3 A0A212F313 A0A1E1WSZ2 I4DQD7 J3JUL2 D6WQ76 N6UI22 A0A1Y1LGS0 A0A1W4WLW4 A0A1W4WBT0 A0A1B6C5U4 A0A0L7LCF7 A0A067RUB6 A0A0T6AZM5 K7IX69 A0A232F9F3 E2C7V6 A0A0J7LBB9 A0A2J7QS09 E1ZY09 A0A026WT89 A0A195FEM0 V5GF95 E0VH70 U5EW28 A0A0K8UWI9 A0A0C9R406 A0A0A1X874 A0A1S3DQX1 A0A0P4VZ98 R4G485 A0A087ZTL5 A0A2A3E6M7 V9IK43 A0A195AWQ8 A0A158NHT7 A0A0K8TRV3 W8ALM3 A0A195EGB3 A0A0P6JZ62 T1DIV1 A0A336LPK7 A0A224XLU4 A0A1B0CI81 A0A1L8EHN0 A0A151WR26 A0A023F9A2 A0A069DPV7 A0A0M5J380 A0A1B6LC06 A0A1D2NIA8 A0A0V0G3N1 A0A0L0CM13 A0A023EID9 T1DF73 A0A1I8MPB5 Q16VD7 A0A0A9ZF23 A0A195CBL1 A0A1L8DV19 A0A3R7PTC9 A0A1Q3EU70 A0A182LI22 Q7PMR4 A0A182YPH5 B4HR19 B4QFD6 F0JAH0 A1Z9M2 Q8T9H5 A0A0L7R0T1 A0A154NYK2 B3MCZ1 A0A0M9A7M6 A0A0P4WZE4 A0A2M4ACF6 Q16VD6 B4P6N0 A0A1W4VDF7 A0A1B6FIY9 A0A2M3Z4D0 B4LLE8 E9HCE1 A0A1Q3EUI2 A0A0P5YI07 A0A0P5Z508 B3NRC2 D3TN14
A0A1E1WM44 A0A2W1BI48 A0A2A4K806 A0A0N1PJ53 S4NXT8 A0A0N1IDF3 A0A212F313 A0A1E1WSZ2 I4DQD7 J3JUL2 D6WQ76 N6UI22 A0A1Y1LGS0 A0A1W4WLW4 A0A1W4WBT0 A0A1B6C5U4 A0A0L7LCF7 A0A067RUB6 A0A0T6AZM5 K7IX69 A0A232F9F3 E2C7V6 A0A0J7LBB9 A0A2J7QS09 E1ZY09 A0A026WT89 A0A195FEM0 V5GF95 E0VH70 U5EW28 A0A0K8UWI9 A0A0C9R406 A0A0A1X874 A0A1S3DQX1 A0A0P4VZ98 R4G485 A0A087ZTL5 A0A2A3E6M7 V9IK43 A0A195AWQ8 A0A158NHT7 A0A0K8TRV3 W8ALM3 A0A195EGB3 A0A0P6JZ62 T1DIV1 A0A336LPK7 A0A224XLU4 A0A1B0CI81 A0A1L8EHN0 A0A151WR26 A0A023F9A2 A0A069DPV7 A0A0M5J380 A0A1B6LC06 A0A1D2NIA8 A0A0V0G3N1 A0A0L0CM13 A0A023EID9 T1DF73 A0A1I8MPB5 Q16VD7 A0A0A9ZF23 A0A195CBL1 A0A1L8DV19 A0A3R7PTC9 A0A1Q3EU70 A0A182LI22 Q7PMR4 A0A182YPH5 B4HR19 B4QFD6 F0JAH0 A1Z9M2 Q8T9H5 A0A0L7R0T1 A0A154NYK2 B3MCZ1 A0A0M9A7M6 A0A0P4WZE4 A0A2M4ACF6 Q16VD6 B4P6N0 A0A1W4VDF7 A0A1B6FIY9 A0A2M3Z4D0 B4LLE8 E9HCE1 A0A1Q3EUI2 A0A0P5YI07 A0A0P5Z508 B3NRC2 D3TN14
Ontologies
PANTHER
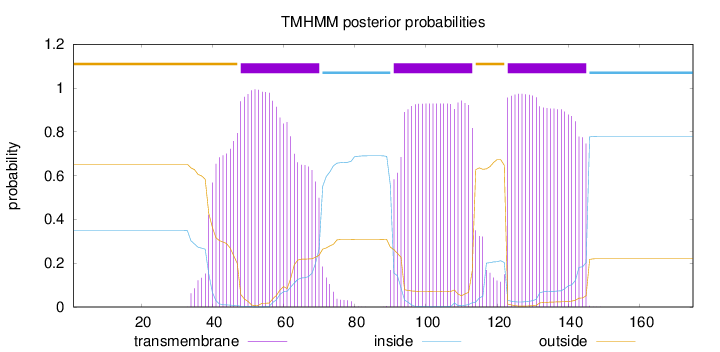
Topology
Subcellular location
Membrane
Length:
175
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
69.28551
Exp number, first 60 AAs:
18.8961
Total prob of N-in:
0.34869
POSSIBLE N-term signal
sequence
outside
1 - 47
TMhelix
48 - 70
inside
71 - 90
TMhelix
91 - 113
outside
114 - 122
TMhelix
123 - 145
inside
146 - 175
Population Genetic Test Statistics
Pi
220.904278
Theta
186.040079
Tajima's D
0.692071
CLR
0.922214
CSRT
0.576271186440678
Interpretation
Uncertain
