Gene
KWMTBOMO16312
Pre Gene Modal
BGIBMGA013774
Annotation
PREDICTED:_uridine_phosphorylase_1-like_isoform_X1_[Bombyx_mori]
Full name
Uridine phosphorylase
Location in the cell
Cytoplasmic Reliability : 3.106
Sequence
CDS
ATGCGTAGAGCTAGATCAAAGTTAGATTCTGTTGCTCTTGAAGAAAGACGCAAAAAGGACCGAGAAAGTAATAAAATGAAGTGTAATTGTGATTACGTTCTTCATTGCAACACTTTGGAGCCGCATTACGACAACTGTGCTACCGTATGGGAGACATCGCACCCAGGAAAGGAATACCCGAAAAACGAAGATGGTACCATCCGCTTGCCAAATAAGAACTTGGATAAGTTGGAATCCGATGTATTGTATCATTTGGGCTTGGATACGAAAAATTACGATCTGCCCGCTTTGTTCGGTGATGTCAAGTTCGTATGTATAGGCGGCACAAAGCATAGAATGAGAGAATTCGCGGAATTTGTTTCATTTAATGTTTTCCATGGAAAAAACATGAAATTGGTTAACCTTACGAAGCATTCGCATCGTTACGCGACTTACAAAGTTGGACCAGTCCTAAGTGTCAGTCACGGGATCGGTATACCGTCGATGACAACGGCCCTACAAGAGATCATTAAGCTGCTGCATTACGCCAAGGCTGTCGACCCTATAGTGTTCAGAATTGGCACGTCGGGGGGGCTCGGTATACCACCAGGTTCCGTCGTTGTATCGAGCTGGGGCCTCAACGGATATTTGGAGAAGAGTTACGATATTCCGGTTATGGGTCGATCATTAAAGCTACCCGCTATTTTAGATAAAAGATTGAGTCAGGAAATATATTCTATGGCATCTGAGGAGGATGAGTTTCAAACATATCTCGGAGGCACGATGGGCGCCGATGATTTTTACAGAGGTCAAGCCCGTTTGGATGGACCTTTCTGTGATCACACAGAAGAGGACAAGCTGGAATTCCTTGAGAAATTATCAGAAATGGGTGTGAGGAATATAGAGATGGAGGCGCCCGCTTTTGCGGCGTTAACAAAAGAAGCCGGCATACGAGGGGCGATCGTTTGTGTTACTCTAATAAACAGACTTGAAGGAGATCAGGTAAGCACCGAGAAGGGTACTTTGTTGCAGTGGCAGAAGAGACCAATGGCTATTGTGGCCAAATTTATATCTAAATACTTAGATAGTATGAACGGATGGAAATAG
Protein
MRRARSKLDSVALEERRKKDRESNKMKCNCDYVLHCNTLEPHYDNCATVWETSHPGKEYPKNEDGTIRLPNKNLDKLESDVLYHLGLDTKNYDLPALFGDVKFVCIGGTKHRMREFAEFVSFNVFHGKNMKLVNLTKHSHRYATYKVGPVLSVSHGIGIPSMTTALQEIIKLLHYAKAVDPIVFRIGTSGGLGIPPGSVVVSSWGLNGYLEKSYDIPVMGRSLKLPAILDKRLSQEIYSMASEEDEFQTYLGGTMGADDFYRGQARLDGPFCDHTEEDKLEFLEKLSEMGVRNIEMEAPAFAALTKEAGIRGAIVCVTLINRLEGDQVSTEKGTLLQWQKRPMAIVAKFISKYLDSMNGWK
Summary
Description
Catalyzes the reversible phosphorylytic cleavage of uridine and deoxyuridine to uracil and ribose- or deoxyribose-1-phosphate. The produced molecules are then utilized as carbon and energy sources or in the rescue of pyrimidine bases for nucleotide synthesis.
Catalytic Activity
phosphate + uridine = alpha-D-ribose 1-phosphate + uracil
Similarity
Belongs to the PNP/UDP phosphorylase family.
Uniprot
H9JW61
A0A2H1VDS7
A0A2A4JF92
A0A2H1VST5
A0A0N1IHY1
A0A0N1IBT0
+ More
A0A3S2M604 N6TC60 A0A1B6E436 A0A232F9X7 V5GTE6 A0A1B6DDL4 V5GHK9 A0A0C9RSZ0 A0A023F5G7 A0A069DYM6 A0A023GIU3 T1HDV8 K7IX68 A0A0T6B3J4 A0A336M4H0 A0A162SIL1 B4JRW5 A0A336KHJ2 A0A0K8TFY7 A0A1S3ID96 A0A2S2QKU5 E9GTM3 A0A1B6KFU3 A0A023EQI1 A0A0A9W2Y6 A0A336KKR0 A0A0P4YSK4 A0A0P5S1P2 A0A0Q9XCR7 A0A067QX58 A0A2R7WS35 A0A2A3E7V8 A0A1I8NAW4 T1PEU7 A0A2J7QZT9 B4K898 A0A146LJI9 A0A1Q3FZS6 A0A154PIC4 A0A084VNU7 A0A182J5Y0 A0A0A9Z8J9 A0A1S4FC12 Q178N8 A0A1Q3G579 A0A146L6D1 A0A182P2A8 A0A2S2NMA6 A0A1S6J0Z4 A0A182N1K9 V9IDR4 A0A0P6I8D6 A0A087ZWP1 Q7PFX7 A0A1E1XIE6 A0A0K8TEY3 A0A182VAL4 A0A182HIB7 A0A182X800 A0A182U4M6 A0A182MVH1 A0A2C9GJV1 A0A0L0C6U7 A0A182QKN0 A0A2A4K1Z2 A0A0Q9WSV4 A0A3B0K7Z1 B4QWT9 B4IC10 A0A2J7QZT7 B4LX04 A0A1B0CE83 A0A182W501 A0A1Y1KPY8 B0X890 Q8IMQ8 A0A0M4F4T1 D6WB44 A0A0Q9WGY3 Q9VBA0 A0A182YQ65 J9K4G6 A0A3B0JLB0 B4NA28 A0A182K465 A0A182R7I4 A0A1L8ED75 A0A212FID5 A0A1I8PMF4 I5AN36 A0A1B6HN66 A0A1B6F7G6 A0A1I8PMD6
A0A3S2M604 N6TC60 A0A1B6E436 A0A232F9X7 V5GTE6 A0A1B6DDL4 V5GHK9 A0A0C9RSZ0 A0A023F5G7 A0A069DYM6 A0A023GIU3 T1HDV8 K7IX68 A0A0T6B3J4 A0A336M4H0 A0A162SIL1 B4JRW5 A0A336KHJ2 A0A0K8TFY7 A0A1S3ID96 A0A2S2QKU5 E9GTM3 A0A1B6KFU3 A0A023EQI1 A0A0A9W2Y6 A0A336KKR0 A0A0P4YSK4 A0A0P5S1P2 A0A0Q9XCR7 A0A067QX58 A0A2R7WS35 A0A2A3E7V8 A0A1I8NAW4 T1PEU7 A0A2J7QZT9 B4K898 A0A146LJI9 A0A1Q3FZS6 A0A154PIC4 A0A084VNU7 A0A182J5Y0 A0A0A9Z8J9 A0A1S4FC12 Q178N8 A0A1Q3G579 A0A146L6D1 A0A182P2A8 A0A2S2NMA6 A0A1S6J0Z4 A0A182N1K9 V9IDR4 A0A0P6I8D6 A0A087ZWP1 Q7PFX7 A0A1E1XIE6 A0A0K8TEY3 A0A182VAL4 A0A182HIB7 A0A182X800 A0A182U4M6 A0A182MVH1 A0A2C9GJV1 A0A0L0C6U7 A0A182QKN0 A0A2A4K1Z2 A0A0Q9WSV4 A0A3B0K7Z1 B4QWT9 B4IC10 A0A2J7QZT7 B4LX04 A0A1B0CE83 A0A182W501 A0A1Y1KPY8 B0X890 Q8IMQ8 A0A0M4F4T1 D6WB44 A0A0Q9WGY3 Q9VBA0 A0A182YQ65 J9K4G6 A0A3B0JLB0 B4NA28 A0A182K465 A0A182R7I4 A0A1L8ED75 A0A212FID5 A0A1I8PMF4 I5AN36 A0A1B6HN66 A0A1B6F7G6 A0A1I8PMD6
EC Number
2.4.2.3
Pubmed
19121390
26354079
23537049
28648823
25474469
26334808
+ More
20075255 17994087 26823975 26383154 21292972 24945155 25401762 24845553 25315136 24438588 17510324 27414796 12364791 14747013 17210077 28503490 26108605 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 25244985 22118469 15632085 23185243
20075255 17994087 26823975 26383154 21292972 24945155 25401762 24845553 25315136 24438588 17510324 27414796 12364791 14747013 17210077 28503490 26108605 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18362917 19820115 25244985 22118469 15632085 23185243
EMBL
BABH01025325
ODYU01001826
SOQ38562.1
NWSH01001676
PCG70446.1
ODYU01004209
+ More
SOQ43836.1 KQ459992 KPJ18578.1 KQ459166 KPJ03577.1 RSAL01000029 RVE51754.1 APGK01043564 KB741015 ENN75323.1 GEDC01022789 GEDC01020497 GEDC01005374 GEDC01004624 JAS14509.1 JAS16801.1 JAS31924.1 JAS32674.1 NNAY01000643 OXU27233.1 GALX01004968 JAB63498.1 GEDC01028640 GEDC01013514 GEDC01006288 GEDC01002289 JAS08658.1 JAS23784.1 JAS31010.1 JAS35009.1 GALX01004967 JAB63499.1 GBYB01011750 JAG81517.1 GBBI01002313 JAC16399.1 GBGD01002125 JAC86764.1 GBBM01001607 JAC33811.1 ACPB03023190 AAZX01000598 LJIG01015983 KRT81967.1 UFQS01000433 UFQT01000433 SSX03876.1 SSX24241.1 LRGB01000024 KZS21337.1 CH916373 EDV94505.1 SSX03877.1 SSX24242.1 GBRD01001776 GDHC01018041 JAG64045.1 JAQ00588.1 GGMS01009141 MBY78344.1 GL732564 EFX77188.1 GEBQ01029650 JAT10327.1 GAPW01001751 JAC11847.1 GBHO01044384 GBHO01037687 JAF99219.1 JAG05917.1 SSX03875.1 SSX24240.1 GDIP01223175 JAJ00227.1 GDIQ01093496 JAL58230.1 CH933806 KRG02161.1 KK852865 KDR14775.1 KK855399 PTY22378.1 KZ288354 PBC27269.1 KA646437 KA646438 AFP61067.1 NEVH01009067 PNF34098.1 EDW16480.1 GDHC01010451 JAQ08178.1 GFDL01001988 JAV33057.1 KQ434924 KZC11562.1 ATLV01014923 KE524996 KFB39641.1 GBHO01037693 GBHO01002022 GBRD01009849 JAG05911.1 JAG41582.1 JAG55975.1 CH477361 EAT42652.1 GFDL01000083 JAV34962.1 GDHC01014625 JAQ04004.1 GGMR01005686 MBY18305.1 KU764443 AQS60689.1 JR038970 AEY58469.1 GDIQ01008074 JAN86663.1 AAAB01008846 EAA45146.4 GFAC01000078 JAT99110.1 GBRD01001777 JAG64044.1 APCN01003925 AXCM01001441 JRES01000835 KNC27961.1 AXCN02002448 NWSH01000241 PCG78079.1 CH964232 KRF99317.1 OUUW01000005 SPP81121.1 CM000364 EDX14606.1 CH480828 EDW45168.1 PNF34097.1 CH940650 EDW67751.1 AJWK01008658 GEZM01080281 JAV62230.1 DS232478 EDS42359.1 AE014297 BT099788 AAN14095.1 ACV91625.1 CP012526 ALC46432.1 KQ971311 EEZ98936.1 KRF83442.1 BT004884 AAF56642.2 AAO45240.1 ABLF02040899 ABLF02040913 SPP81122.1 EDW80671.1 GFDG01002109 JAV16690.1 AGBW02008396 OWR53503.1 CM000070 EIM52371.1 KRS99772.1 GECU01031587 JAS76119.1 GECZ01023698 JAS46071.1
SOQ43836.1 KQ459992 KPJ18578.1 KQ459166 KPJ03577.1 RSAL01000029 RVE51754.1 APGK01043564 KB741015 ENN75323.1 GEDC01022789 GEDC01020497 GEDC01005374 GEDC01004624 JAS14509.1 JAS16801.1 JAS31924.1 JAS32674.1 NNAY01000643 OXU27233.1 GALX01004968 JAB63498.1 GEDC01028640 GEDC01013514 GEDC01006288 GEDC01002289 JAS08658.1 JAS23784.1 JAS31010.1 JAS35009.1 GALX01004967 JAB63499.1 GBYB01011750 JAG81517.1 GBBI01002313 JAC16399.1 GBGD01002125 JAC86764.1 GBBM01001607 JAC33811.1 ACPB03023190 AAZX01000598 LJIG01015983 KRT81967.1 UFQS01000433 UFQT01000433 SSX03876.1 SSX24241.1 LRGB01000024 KZS21337.1 CH916373 EDV94505.1 SSX03877.1 SSX24242.1 GBRD01001776 GDHC01018041 JAG64045.1 JAQ00588.1 GGMS01009141 MBY78344.1 GL732564 EFX77188.1 GEBQ01029650 JAT10327.1 GAPW01001751 JAC11847.1 GBHO01044384 GBHO01037687 JAF99219.1 JAG05917.1 SSX03875.1 SSX24240.1 GDIP01223175 JAJ00227.1 GDIQ01093496 JAL58230.1 CH933806 KRG02161.1 KK852865 KDR14775.1 KK855399 PTY22378.1 KZ288354 PBC27269.1 KA646437 KA646438 AFP61067.1 NEVH01009067 PNF34098.1 EDW16480.1 GDHC01010451 JAQ08178.1 GFDL01001988 JAV33057.1 KQ434924 KZC11562.1 ATLV01014923 KE524996 KFB39641.1 GBHO01037693 GBHO01002022 GBRD01009849 JAG05911.1 JAG41582.1 JAG55975.1 CH477361 EAT42652.1 GFDL01000083 JAV34962.1 GDHC01014625 JAQ04004.1 GGMR01005686 MBY18305.1 KU764443 AQS60689.1 JR038970 AEY58469.1 GDIQ01008074 JAN86663.1 AAAB01008846 EAA45146.4 GFAC01000078 JAT99110.1 GBRD01001777 JAG64044.1 APCN01003925 AXCM01001441 JRES01000835 KNC27961.1 AXCN02002448 NWSH01000241 PCG78079.1 CH964232 KRF99317.1 OUUW01000005 SPP81121.1 CM000364 EDX14606.1 CH480828 EDW45168.1 PNF34097.1 CH940650 EDW67751.1 AJWK01008658 GEZM01080281 JAV62230.1 DS232478 EDS42359.1 AE014297 BT099788 AAN14095.1 ACV91625.1 CP012526 ALC46432.1 KQ971311 EEZ98936.1 KRF83442.1 BT004884 AAF56642.2 AAO45240.1 ABLF02040899 ABLF02040913 SPP81122.1 EDW80671.1 GFDG01002109 JAV16690.1 AGBW02008396 OWR53503.1 CM000070 EIM52371.1 KRS99772.1 GECU01031587 JAS76119.1 GECZ01023698 JAS46071.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000019118
+ More
UP000215335 UP000015103 UP000002358 UP000076858 UP000001070 UP000085678 UP000000305 UP000009192 UP000027135 UP000242457 UP000095301 UP000235965 UP000076502 UP000030765 UP000075880 UP000008820 UP000075885 UP000075884 UP000005203 UP000007062 UP000075903 UP000075840 UP000076407 UP000075902 UP000075883 UP000037069 UP000075886 UP000007798 UP000268350 UP000000304 UP000001292 UP000008792 UP000092461 UP000075920 UP000002320 UP000000803 UP000092553 UP000007266 UP000076408 UP000007819 UP000075881 UP000075900 UP000007151 UP000095300 UP000001819
UP000215335 UP000015103 UP000002358 UP000076858 UP000001070 UP000085678 UP000000305 UP000009192 UP000027135 UP000242457 UP000095301 UP000235965 UP000076502 UP000030765 UP000075880 UP000008820 UP000075885 UP000075884 UP000005203 UP000007062 UP000075903 UP000075840 UP000076407 UP000075902 UP000075883 UP000037069 UP000075886 UP000007798 UP000268350 UP000000304 UP000001292 UP000008792 UP000092461 UP000075920 UP000002320 UP000000803 UP000092553 UP000007266 UP000076408 UP000007819 UP000075881 UP000075900 UP000007151 UP000095300 UP000001819
Pfam
PF01048 PNP_UDP_1
Interpro
SUPFAM
SSF53167
SSF53167
Gene 3D
ProteinModelPortal
H9JW61
A0A2H1VDS7
A0A2A4JF92
A0A2H1VST5
A0A0N1IHY1
A0A0N1IBT0
+ More
A0A3S2M604 N6TC60 A0A1B6E436 A0A232F9X7 V5GTE6 A0A1B6DDL4 V5GHK9 A0A0C9RSZ0 A0A023F5G7 A0A069DYM6 A0A023GIU3 T1HDV8 K7IX68 A0A0T6B3J4 A0A336M4H0 A0A162SIL1 B4JRW5 A0A336KHJ2 A0A0K8TFY7 A0A1S3ID96 A0A2S2QKU5 E9GTM3 A0A1B6KFU3 A0A023EQI1 A0A0A9W2Y6 A0A336KKR0 A0A0P4YSK4 A0A0P5S1P2 A0A0Q9XCR7 A0A067QX58 A0A2R7WS35 A0A2A3E7V8 A0A1I8NAW4 T1PEU7 A0A2J7QZT9 B4K898 A0A146LJI9 A0A1Q3FZS6 A0A154PIC4 A0A084VNU7 A0A182J5Y0 A0A0A9Z8J9 A0A1S4FC12 Q178N8 A0A1Q3G579 A0A146L6D1 A0A182P2A8 A0A2S2NMA6 A0A1S6J0Z4 A0A182N1K9 V9IDR4 A0A0P6I8D6 A0A087ZWP1 Q7PFX7 A0A1E1XIE6 A0A0K8TEY3 A0A182VAL4 A0A182HIB7 A0A182X800 A0A182U4M6 A0A182MVH1 A0A2C9GJV1 A0A0L0C6U7 A0A182QKN0 A0A2A4K1Z2 A0A0Q9WSV4 A0A3B0K7Z1 B4QWT9 B4IC10 A0A2J7QZT7 B4LX04 A0A1B0CE83 A0A182W501 A0A1Y1KPY8 B0X890 Q8IMQ8 A0A0M4F4T1 D6WB44 A0A0Q9WGY3 Q9VBA0 A0A182YQ65 J9K4G6 A0A3B0JLB0 B4NA28 A0A182K465 A0A182R7I4 A0A1L8ED75 A0A212FID5 A0A1I8PMF4 I5AN36 A0A1B6HN66 A0A1B6F7G6 A0A1I8PMD6
A0A3S2M604 N6TC60 A0A1B6E436 A0A232F9X7 V5GTE6 A0A1B6DDL4 V5GHK9 A0A0C9RSZ0 A0A023F5G7 A0A069DYM6 A0A023GIU3 T1HDV8 K7IX68 A0A0T6B3J4 A0A336M4H0 A0A162SIL1 B4JRW5 A0A336KHJ2 A0A0K8TFY7 A0A1S3ID96 A0A2S2QKU5 E9GTM3 A0A1B6KFU3 A0A023EQI1 A0A0A9W2Y6 A0A336KKR0 A0A0P4YSK4 A0A0P5S1P2 A0A0Q9XCR7 A0A067QX58 A0A2R7WS35 A0A2A3E7V8 A0A1I8NAW4 T1PEU7 A0A2J7QZT9 B4K898 A0A146LJI9 A0A1Q3FZS6 A0A154PIC4 A0A084VNU7 A0A182J5Y0 A0A0A9Z8J9 A0A1S4FC12 Q178N8 A0A1Q3G579 A0A146L6D1 A0A182P2A8 A0A2S2NMA6 A0A1S6J0Z4 A0A182N1K9 V9IDR4 A0A0P6I8D6 A0A087ZWP1 Q7PFX7 A0A1E1XIE6 A0A0K8TEY3 A0A182VAL4 A0A182HIB7 A0A182X800 A0A182U4M6 A0A182MVH1 A0A2C9GJV1 A0A0L0C6U7 A0A182QKN0 A0A2A4K1Z2 A0A0Q9WSV4 A0A3B0K7Z1 B4QWT9 B4IC10 A0A2J7QZT7 B4LX04 A0A1B0CE83 A0A182W501 A0A1Y1KPY8 B0X890 Q8IMQ8 A0A0M4F4T1 D6WB44 A0A0Q9WGY3 Q9VBA0 A0A182YQ65 J9K4G6 A0A3B0JLB0 B4NA28 A0A182K465 A0A182R7I4 A0A1L8ED75 A0A212FID5 A0A1I8PMF4 I5AN36 A0A1B6HN66 A0A1B6F7G6 A0A1I8PMD6
PDB
3NBQ
E-value=3.81214e-75,
Score=715
Ontologies
PATHWAY
GO
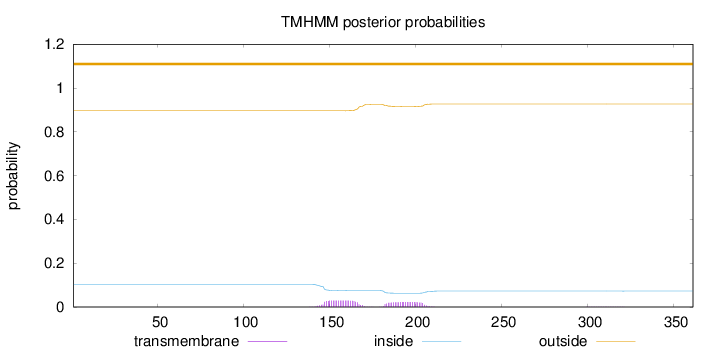
Topology
Length:
361
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.14148
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10416
outside
1 - 361
Population Genetic Test Statistics
Pi
235.901952
Theta
188.584136
Tajima's D
-2.654927
CLR
164.93995
CSRT
0.000149992500374981
Interpretation
Possibly Positive selection
