Gene
KWMTBOMO16311
Pre Gene Modal
BGIBMGA013766
Annotation
PREDICTED:_stabilizer_of_axonemal_microtubules_1_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 1.723 Nuclear Reliability : 2.083
Sequence
CDS
ATGAGACCGAACTGCCCATGCCGATCTTGCTGCTGTGGGAAACCGCCCATTGTTAAGCCGTGTTACAAGCAACCAAAAATCCCAGAGTCGTACGCGCCGAGACGATGCTACATAAAACCATCAGCCCCCGTAGAGGGATGCACGACTTACAAGCTGTCGTATCTACCTGTCGACGGATGCAAAAACCTACGGGGAGAAGTAAAGAAACCTTCGCCCAACATTGTACCGAGCTGTGAACCTATGGAAGGGTGTACCGTTCAAAAGTTGTCATACCTACCGAACCCGGTGTGCGTGACGCAGTCGATCCGGCCCTGCCACCACGACATGTGGGGCCAGGGTCCCATGCAGAACATCACCACACAACGACACGACTACGTGCCCAAGCCGAGTATACTACGAGAGAGCTTTAAGCCAGCACCGAAATTTCACTGCGTCGACCAACCATTTGAAAATCGGACAATAAATAAATTATCTTATTTACCGCCCGAGAAAATTGATATGCCGAAATCGTTTGCGCCCGAGCGTTGCTATGAAAAGCCGGCTGCCAAAATGGACGGAACCACCACCCACAAGATGAGCTATTTACCGAATCCTGTAATGAAGAAAGAAACAGTACCGTGGGCTTGTAAGGGACAATATGAGAAGCCTTGTCAAAAGTTGGACGGAAATACGACATACACAATGAGTTACTTGGACGCGAAAAGCGACTGTCGTCGACAACCCATCTTACCGACTGCTTGCTCCAACCCTGTCTCCGCTTCGAAGAGATTTGAAACGCAGACTATTTATAAGAACAGCTATTAA
Protein
MRPNCPCRSCCCGKPPIVKPCYKQPKIPESYAPRRCYIKPSAPVEGCTTYKLSYLPVDGCKNLRGEVKKPSPNIVPSCEPMEGCTVQKLSYLPNPVCVTQSIRPCHHDMWGQGPMQNITTQRHDYVPKPSILRESFKPAPKFHCVDQPFENRTINKLSYLPPEKIDMPKSFAPERCYEKPAAKMDGTTTHKMSYLPNPVMKKETVPWACKGQYEKPCQKLDGNTTYTMSYLDAKSDCRRQPILPTACSNPVSASKRFETQTIYKNSY
Summary
Uniprot
A0A2H1VGE3
A0A2A4JEE8
A0A2H1VIJ2
A0A212FEK3
A0A3S2TPP0
A0A2W1BCV8
+ More
A0A0N0P9T8 A0A0N0PEC1 A0A0L7L3N2 D6WXL0 N6UCZ6 A0A182PFG4 T1HSU0 A0A1W4X5U4 E2BNI7 A0A084W4Y5 A0A1A9VVU6 A0A182N6Q4 A0A1B0FLE3 E2BN65 A0A170Z048 A0A1A9Z3Q2 A0A182UP61 A0A2A3ECD5 Q7QBL0 A0A182XEB1 A0A182HJ11 A0A182U1F5 A0A182RYE4 A0A1I8MRZ6 A0A088AFG3 A0A182Q271 A0A182W2V2 A0A0L7QKX8 A0A182LC67 A0A1S4F7F2 A0A182XVL8 A0A0M4EEX8 A0A023EWG9 A0A151J364 A0A182G5I2 A0A1Q3G027 A0A151J7D1 A0A1Q3FZX3 A0A1A9YLZ3 Q9VE97 A0A1A9WRV5 A0A151WVL1 A0A1Y1LH75 A0A158NFS0 B3NZ77 A0A0N0BL28 A0A1A9Y401 A0A182MNJ0 A0A1B0BSN9 A0A182FAA7 A0A1W4U8L8 B4IB69 B4GMF2 Q294X4 B4NG79 A0A182IXY2 A0A0A9XKX1 A0A3B0K2C0 A0A1I8PW92 A0A3L8E022 A0A026WAF5 B4QUG5 A0A034VW42 A0A0K8U611 B3MTP7 W8BZH5 A0A158NU77 B4LZA0 A0A1B6DE04 A0A026WRX9 E2AP85 A0A310SCM6 K7J0E5 W5J615 B4JZ21 A0A088AM40 B4KD75 A0A0L0BWB7 A0A023ELF7 A0A1B6LFP9 A0A0J7L7A5 E9IHK0 F4WR55 A0A1B6FNK7 A0A232FBM2 A0A151WET0 T1GL73 E0VSG8 A0A195D702
A0A0N0P9T8 A0A0N0PEC1 A0A0L7L3N2 D6WXL0 N6UCZ6 A0A182PFG4 T1HSU0 A0A1W4X5U4 E2BNI7 A0A084W4Y5 A0A1A9VVU6 A0A182N6Q4 A0A1B0FLE3 E2BN65 A0A170Z048 A0A1A9Z3Q2 A0A182UP61 A0A2A3ECD5 Q7QBL0 A0A182XEB1 A0A182HJ11 A0A182U1F5 A0A182RYE4 A0A1I8MRZ6 A0A088AFG3 A0A182Q271 A0A182W2V2 A0A0L7QKX8 A0A182LC67 A0A1S4F7F2 A0A182XVL8 A0A0M4EEX8 A0A023EWG9 A0A151J364 A0A182G5I2 A0A1Q3G027 A0A151J7D1 A0A1Q3FZX3 A0A1A9YLZ3 Q9VE97 A0A1A9WRV5 A0A151WVL1 A0A1Y1LH75 A0A158NFS0 B3NZ77 A0A0N0BL28 A0A1A9Y401 A0A182MNJ0 A0A1B0BSN9 A0A182FAA7 A0A1W4U8L8 B4IB69 B4GMF2 Q294X4 B4NG79 A0A182IXY2 A0A0A9XKX1 A0A3B0K2C0 A0A1I8PW92 A0A3L8E022 A0A026WAF5 B4QUG5 A0A034VW42 A0A0K8U611 B3MTP7 W8BZH5 A0A158NU77 B4LZA0 A0A1B6DE04 A0A026WRX9 E2AP85 A0A310SCM6 K7J0E5 W5J615 B4JZ21 A0A088AM40 B4KD75 A0A0L0BWB7 A0A023ELF7 A0A1B6LFP9 A0A0J7L7A5 E9IHK0 F4WR55 A0A1B6FNK7 A0A232FBM2 A0A151WET0 T1GL73 E0VSG8 A0A195D702
Pubmed
22118469
28756777
26354079
26227816
18362917
19820115
+ More
23537049 20798317 24438588 12364791 14747013 17210077 25315136 20966253 25244985 24945155 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28004739 21347285 17994087 15632085 23185243 25401762 30249741 24508170 25348373 24495485 20075255 20920257 23761445 18057021 26108605 21282665 21719571 28648823 20566863
23537049 20798317 24438588 12364791 14747013 17210077 25315136 20966253 25244985 24945155 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28004739 21347285 17994087 15632085 23185243 25401762 30249741 24508170 25348373 24495485 20075255 20920257 23761445 18057021 26108605 21282665 21719571 28648823 20566863
EMBL
ODYU01002424
SOQ39887.1
NWSH01001676
PCG70447.1
ODYU01002757
SOQ40648.1
+ More
AGBW02008924 OWR52179.1 RSAL01000029 RVE51753.1 KZ150264 PZC71715.1 KQ459166 KPJ03578.1 KQ459992 KPJ18575.1 JTDY01003127 KOB70103.1 KQ971361 EFA07966.1 APGK01040034 KB740975 KB631935 ENN76522.1 ERL87329.1 ACPB03016664 GL449414 EFN82766.1 ATLV01020424 KE525302 KFB45279.1 CCAG010010321 GL449385 EFN82827.1 GEMB01002766 JAS00431.1 KZ288287 PBC29397.1 AAAB01008879 EAA08491.5 APCN01002114 AXCN02000061 KQ414940 KOC59181.1 CP012526 ALC46628.1 GAPW01000784 JAC12814.1 KQ980298 KYN16843.1 JXUM01146027 JXUM01146028 JXUM01146029 JXUM01146030 KQ570097 KXJ68458.1 GFDL01001929 JAV33116.1 KQ979748 KYN19354.1 GFDL01001927 JAV33118.1 AE014297 AY119515 AAF55530.2 AAM50169.1 KQ982699 KYQ51942.1 GEZM01057613 JAV72218.1 ADTU01014656 CH954181 EDV48619.1 KQ435689 KOX81291.1 AXCM01000405 JXJN01019782 JXJN01019783 CH480827 EDW44627.1 CH479185 EDW38026.1 CM000070 EAL28838.2 KRT01106.1 CH964251 EDW83296.1 GBHO01023015 GBRD01003794 JAG20589.1 JAG62027.1 OUUW01000014 SPP88445.1 QOIP01000002 RLU25735.1 KK107323 EZA52606.1 CM000364 EDX12465.1 GAKP01011391 JAC47561.1 GDHF01030225 JAI22089.1 CH902623 EDV30178.1 GAMC01007839 JAB98716.1 ADTU01026284 ADTU01026285 CH940650 EDW68135.1 GEDC01013438 JAS23860.1 KK107119 EZA58805.1 GL441481 EFN64758.1 KQ777939 OAD52033.1 ADMH02002087 ETN59411.1 CH916378 EDV98636.1 CH933806 EDW14857.1 KRG01247.1 JRES01001243 KNC24322.1 GAPW01003546 JAC10052.1 GEBQ01017469 JAT22508.1 LBMM01000419 KMQ98409.1 GL763289 EFZ19924.1 GL888284 EGI63282.1 GECZ01018176 JAS51593.1 NNAY01000459 OXU28244.1 KQ983238 KYQ46353.1 CAQQ02129165 CAQQ02129166 DS235751 EEB16324.1 KQ976750 KYN08653.1
AGBW02008924 OWR52179.1 RSAL01000029 RVE51753.1 KZ150264 PZC71715.1 KQ459166 KPJ03578.1 KQ459992 KPJ18575.1 JTDY01003127 KOB70103.1 KQ971361 EFA07966.1 APGK01040034 KB740975 KB631935 ENN76522.1 ERL87329.1 ACPB03016664 GL449414 EFN82766.1 ATLV01020424 KE525302 KFB45279.1 CCAG010010321 GL449385 EFN82827.1 GEMB01002766 JAS00431.1 KZ288287 PBC29397.1 AAAB01008879 EAA08491.5 APCN01002114 AXCN02000061 KQ414940 KOC59181.1 CP012526 ALC46628.1 GAPW01000784 JAC12814.1 KQ980298 KYN16843.1 JXUM01146027 JXUM01146028 JXUM01146029 JXUM01146030 KQ570097 KXJ68458.1 GFDL01001929 JAV33116.1 KQ979748 KYN19354.1 GFDL01001927 JAV33118.1 AE014297 AY119515 AAF55530.2 AAM50169.1 KQ982699 KYQ51942.1 GEZM01057613 JAV72218.1 ADTU01014656 CH954181 EDV48619.1 KQ435689 KOX81291.1 AXCM01000405 JXJN01019782 JXJN01019783 CH480827 EDW44627.1 CH479185 EDW38026.1 CM000070 EAL28838.2 KRT01106.1 CH964251 EDW83296.1 GBHO01023015 GBRD01003794 JAG20589.1 JAG62027.1 OUUW01000014 SPP88445.1 QOIP01000002 RLU25735.1 KK107323 EZA52606.1 CM000364 EDX12465.1 GAKP01011391 JAC47561.1 GDHF01030225 JAI22089.1 CH902623 EDV30178.1 GAMC01007839 JAB98716.1 ADTU01026284 ADTU01026285 CH940650 EDW68135.1 GEDC01013438 JAS23860.1 KK107119 EZA58805.1 GL441481 EFN64758.1 KQ777939 OAD52033.1 ADMH02002087 ETN59411.1 CH916378 EDV98636.1 CH933806 EDW14857.1 KRG01247.1 JRES01001243 KNC24322.1 GAPW01003546 JAC10052.1 GEBQ01017469 JAT22508.1 LBMM01000419 KMQ98409.1 GL763289 EFZ19924.1 GL888284 EGI63282.1 GECZ01018176 JAS51593.1 NNAY01000459 OXU28244.1 KQ983238 KYQ46353.1 CAQQ02129165 CAQQ02129166 DS235751 EEB16324.1 KQ976750 KYN08653.1
Proteomes
UP000218220
UP000007151
UP000283053
UP000053268
UP000053240
UP000037510
+ More
UP000007266 UP000019118 UP000030742 UP000075885 UP000015103 UP000192223 UP000008237 UP000030765 UP000078200 UP000075884 UP000092444 UP000092445 UP000075903 UP000242457 UP000007062 UP000076407 UP000075840 UP000075902 UP000075900 UP000095301 UP000005203 UP000075886 UP000075920 UP000053825 UP000075882 UP000076408 UP000092553 UP000078492 UP000069940 UP000249989 UP000092443 UP000000803 UP000091820 UP000075809 UP000005205 UP000008711 UP000053105 UP000075883 UP000092460 UP000069272 UP000192221 UP000001292 UP000008744 UP000001819 UP000007798 UP000075880 UP000268350 UP000095300 UP000279307 UP000053097 UP000000304 UP000007801 UP000008792 UP000000311 UP000002358 UP000000673 UP000001070 UP000009192 UP000037069 UP000036403 UP000007755 UP000215335 UP000015102 UP000009046 UP000078542
UP000007266 UP000019118 UP000030742 UP000075885 UP000015103 UP000192223 UP000008237 UP000030765 UP000078200 UP000075884 UP000092444 UP000092445 UP000075903 UP000242457 UP000007062 UP000076407 UP000075840 UP000075902 UP000075900 UP000095301 UP000005203 UP000075886 UP000075920 UP000053825 UP000075882 UP000076408 UP000092553 UP000078492 UP000069940 UP000249989 UP000092443 UP000000803 UP000091820 UP000075809 UP000005205 UP000008711 UP000053105 UP000075883 UP000092460 UP000069272 UP000192221 UP000001292 UP000008744 UP000001819 UP000007798 UP000075880 UP000268350 UP000095300 UP000279307 UP000053097 UP000000304 UP000007801 UP000008792 UP000000311 UP000002358 UP000000673 UP000001070 UP000009192 UP000037069 UP000036403 UP000007755 UP000215335 UP000015102 UP000009046 UP000078542
PRIDE
Pfam
Interpro
IPR033336
SAXO1/2
+ More
IPR000403 PI3/4_kinase_cat_dom
IPR039756 Lsb6/PI4K2
IPR000618 Insect_cuticle
IPR031311 CHIT_BIND_RR_consensus
IPR011990 TPR-like_helical_dom_sf
IPR037387 PTCD3
IPR033443 PPR_long
IPR002885 Pentatricopeptide_repeat
IPR008971 HSP40/DnaJ_pept-bd
IPR028085 FNIP_mid_dom
IPR028084 FNIP_N_dom
IPR037545 DENN_FNIP1/2
IPR002939 DnaJ_C
IPR026156 FNIP_fam
IPR028086 FNIP_C_dom
IPR000403 PI3/4_kinase_cat_dom
IPR039756 Lsb6/PI4K2
IPR000618 Insect_cuticle
IPR031311 CHIT_BIND_RR_consensus
IPR011990 TPR-like_helical_dom_sf
IPR037387 PTCD3
IPR033443 PPR_long
IPR002885 Pentatricopeptide_repeat
IPR008971 HSP40/DnaJ_pept-bd
IPR028085 FNIP_mid_dom
IPR028084 FNIP_N_dom
IPR037545 DENN_FNIP1/2
IPR002939 DnaJ_C
IPR026156 FNIP_fam
IPR028086 FNIP_C_dom
SUPFAM
SSF49493
SSF49493
Gene 3D
ProteinModelPortal
A0A2H1VGE3
A0A2A4JEE8
A0A2H1VIJ2
A0A212FEK3
A0A3S2TPP0
A0A2W1BCV8
+ More
A0A0N0P9T8 A0A0N0PEC1 A0A0L7L3N2 D6WXL0 N6UCZ6 A0A182PFG4 T1HSU0 A0A1W4X5U4 E2BNI7 A0A084W4Y5 A0A1A9VVU6 A0A182N6Q4 A0A1B0FLE3 E2BN65 A0A170Z048 A0A1A9Z3Q2 A0A182UP61 A0A2A3ECD5 Q7QBL0 A0A182XEB1 A0A182HJ11 A0A182U1F5 A0A182RYE4 A0A1I8MRZ6 A0A088AFG3 A0A182Q271 A0A182W2V2 A0A0L7QKX8 A0A182LC67 A0A1S4F7F2 A0A182XVL8 A0A0M4EEX8 A0A023EWG9 A0A151J364 A0A182G5I2 A0A1Q3G027 A0A151J7D1 A0A1Q3FZX3 A0A1A9YLZ3 Q9VE97 A0A1A9WRV5 A0A151WVL1 A0A1Y1LH75 A0A158NFS0 B3NZ77 A0A0N0BL28 A0A1A9Y401 A0A182MNJ0 A0A1B0BSN9 A0A182FAA7 A0A1W4U8L8 B4IB69 B4GMF2 Q294X4 B4NG79 A0A182IXY2 A0A0A9XKX1 A0A3B0K2C0 A0A1I8PW92 A0A3L8E022 A0A026WAF5 B4QUG5 A0A034VW42 A0A0K8U611 B3MTP7 W8BZH5 A0A158NU77 B4LZA0 A0A1B6DE04 A0A026WRX9 E2AP85 A0A310SCM6 K7J0E5 W5J615 B4JZ21 A0A088AM40 B4KD75 A0A0L0BWB7 A0A023ELF7 A0A1B6LFP9 A0A0J7L7A5 E9IHK0 F4WR55 A0A1B6FNK7 A0A232FBM2 A0A151WET0 T1GL73 E0VSG8 A0A195D702
A0A0N0P9T8 A0A0N0PEC1 A0A0L7L3N2 D6WXL0 N6UCZ6 A0A182PFG4 T1HSU0 A0A1W4X5U4 E2BNI7 A0A084W4Y5 A0A1A9VVU6 A0A182N6Q4 A0A1B0FLE3 E2BN65 A0A170Z048 A0A1A9Z3Q2 A0A182UP61 A0A2A3ECD5 Q7QBL0 A0A182XEB1 A0A182HJ11 A0A182U1F5 A0A182RYE4 A0A1I8MRZ6 A0A088AFG3 A0A182Q271 A0A182W2V2 A0A0L7QKX8 A0A182LC67 A0A1S4F7F2 A0A182XVL8 A0A0M4EEX8 A0A023EWG9 A0A151J364 A0A182G5I2 A0A1Q3G027 A0A151J7D1 A0A1Q3FZX3 A0A1A9YLZ3 Q9VE97 A0A1A9WRV5 A0A151WVL1 A0A1Y1LH75 A0A158NFS0 B3NZ77 A0A0N0BL28 A0A1A9Y401 A0A182MNJ0 A0A1B0BSN9 A0A182FAA7 A0A1W4U8L8 B4IB69 B4GMF2 Q294X4 B4NG79 A0A182IXY2 A0A0A9XKX1 A0A3B0K2C0 A0A1I8PW92 A0A3L8E022 A0A026WAF5 B4QUG5 A0A034VW42 A0A0K8U611 B3MTP7 W8BZH5 A0A158NU77 B4LZA0 A0A1B6DE04 A0A026WRX9 E2AP85 A0A310SCM6 K7J0E5 W5J615 B4JZ21 A0A088AM40 B4KD75 A0A0L0BWB7 A0A023ELF7 A0A1B6LFP9 A0A0J7L7A5 E9IHK0 F4WR55 A0A1B6FNK7 A0A232FBM2 A0A151WET0 T1GL73 E0VSG8 A0A195D702
Ontologies
GO
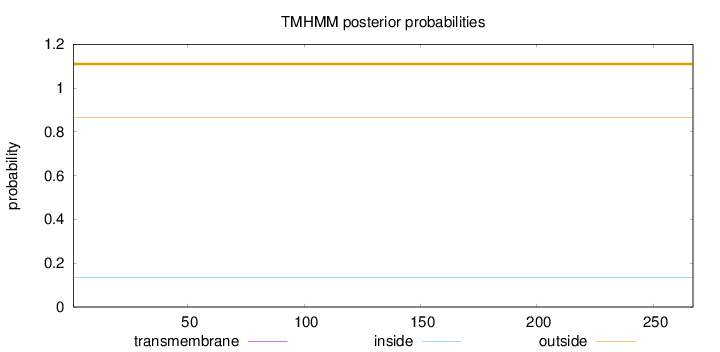
Topology
Length:
267
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.13608
outside
1 - 267
Population Genetic Test Statistics
Pi
178.522545
Theta
154.447064
Tajima's D
-1.369414
CLR
2026.684044
CSRT
0.080345982700865
Interpretation
Possibly Positive selection
