Pre Gene Modal
BGIBMGA013773
Annotation
PREDICTED:_NSFL1_cofactor_p47-like_[Bombyx_mori]
Full name
NSFL1 cofactor p47
Alternative Name
p97 cofactor p47
Location in the cell
Nuclear Reliability : 3.471
Sequence
CDS
ATGTCTGCGAATAGAGAAGATACATTACGTCAGTTTTGTGATGTTACCGGTGCTGACGAAGATCGCAGTAAATTCTTTTTAGAATCCTCAAACTGGCAATTAGATGTTGCTCTTTCAAGTTTCTATGAGAACGGTGGTAATGCAGATGAGGCTCCAGCTAATCCAACGAGTGCGGCATCATTCTCTGTTCTGCCAGACAGCGATATGGAATCACCGCCCGTCTCACCTCAGGTACAAAAGTCCCAGAAAAAAGAAAAAAAGAAAACCTCAAATACAAAGTTTGGGACCATAGGATCGCTGCAGCAGGAAAGCTCAAGCGATGAGGAAGAAGGTCAGGCATTTTATGCTGGGGGCTCAGAACGGTCCGGGCAACAGATCTTAGGGCCAAGTAAAGGCAGGAAAGATATCGTCTCTGAAATGTTCAAGAGCGTCAGAGAGCGAGGAGCAGTCGTCTTTGAAGACGAACCAACATCCAGTAGAGGCCGTGGTCGAGGGGGCCTCTTCGCCGGAGTTGGCTATAGATTAGGACAAACTGCAGATGATCACGAGCCAGTGTCTGCATCGAATCCACAACAACAAGAGCAAGTGAGACTGGTGAGACTGTGTCTGTACCGCGAAGGCTTCACGGTGGACGCGGGTCCGCTGCGCCACTACAGCGACCCGGAGAACGCGGAGTTCCTGAGCTGCATACGTCGCGGTGAGATTCCGTCGGAGTTGTCGGGGCAGGGGGTCTCGGTCCGGGTGAGCCTCGAGGACAGCAGACACGAGGAGTGTCCCCGGCACGTGGTCAAGCCGCAGGCCTTCGCCGGGAAAGGGCACATGCTGGGCAGTCCCACGCCCCCCACGGTGGGCGCCACAGCCGCCGTGGCGGGCGACCAGGCCGACCGCGCGGCCAACCTCCGCGCCGCACAGAGCGCCGTCACACTCGACGAAACGCAACCCGTCACTACGCTGCAGTTCCGACTGGCCGACGGCACCCGCATCACGGGCCGGTTCAACCACAGCCACACGGTCGGCGCGCTGAGGGAGTTCGTGTCGCGAGCGGAGCCGGCGTACCAGCTGCGGCCATTCAGTCTGCTGACGGCCTTCCCGAGCGCGGAGCTGGGTGACGACGCCGCGACCCTGGCGCAGGCGAACCTGCTCAACTCGACGGTCATGCAGCGGCTCAAGTAG
Protein
MSANREDTLRQFCDVTGADEDRSKFFLESSNWQLDVALSSFYENGGNADEAPANPTSAASFSVLPDSDMESPPVSPQVQKSQKKEKKKTSNTKFGTIGSLQQESSSDEEEGQAFYAGGSERSGQQILGPSKGRKDIVSEMFKSVRERGAVVFEDEPTSSRGRGRGGLFAGVGYRLGQTADDHEPVSASNPQQQEQVRLVRLCLYREGFTVDAGPLRHYSDPENAEFLSCIRRGEIPSELSGQGVSVRVSLEDSRHEECPRHVVKPQAFAGKGHMLGSPTPPTVGATAAVAGDQADRAANLRAAQSAVTLDETQPVTTLQFRLADGTRITGRFNHSHTVGALREFVSRAEPAYQLRPFSLLTAFPSAELGDDAATLAQANLLNSTVMQRLK
Summary
Description
Reduces the ATPase activity of VCP. Necessary for the fragmentation of Golgi stacks during mitosis and for VCP-mediated reassembly of Golgi stacks after mitosis. May play a role in VCP-mediated formation of transitional endoplasmic reticulum (tER). Inhibits the activity of CTSL (in vitro). Together with UBXN2B/p37, regulates the centrosomal levels of kinase AURKA/Aurora A levels during mitotic progression by promoting AURKA removal from centrosomes in prophase. Also, regulates spindle orientation during mitosis.
Similarity
Belongs to the NSFL1C family.
Keywords
Complete proteome
Cytoplasm
Cytoskeleton
Golgi apparatus
Lipid-binding
Nucleus
Reference proteome
Feature
chain NSFL1 cofactor p47
Uniprot
A0A2A4J2R8
A0A3S2P3Y6
S4PY40
A0A2H1VIJ3
A0A2A4J3Y5
A0A1E1WA73
+ More
A0A2W1BD25 A0A1B6KXR7 A0A2J7PD98 A0A2J7PDA4 A0A1Y1L4L0 D6WT40 A0A151IV27 A0A3L8DFP9 A0A195F004 E2AKL7 F4WAT6 A0A151XDM5 A0A195BWY2 A0A151IH76 A0A158P3K4 E9J8R6 A0A182GLM9 Q1HRG9 A0A0J7LAA6 A0A087ZQ92 A0A162D7N3 A0A232FC07 A0A1W4WTH1 A0A2A3EJ17 V9IDV9 A0A310S969 E2BNT9 K7J497 A0A0M9A2E6 E9FYH5 U5EYU9 A0A0K8TSI9 A0A0P4W4J8 A0A0P5CXR9 A0A182FH85 A0A154PIP1 V5H138 A0A1I8M796 T1PDZ5 A0A2Y9F8U5 A0A2Y9PSG9 A0A341B7H5 A0A2U3V8Q1 A0A084W9K5 A0A2M4BQM0 A0A2M4ADL1 A0A067R7M4 A0A2M4BQE1 A0A2Y9PTH9 A0A0L7R492 A0A2Y9F790 A0A341BAU6 A0A2U3V8B0 A0A1S2ZQU1 A0A2U4CLV0 A0A341BAU0 A0A2Y9PYU9 A0A340Y1D4 C3Y288 W5JP15 A0A3Q2XMI0 A0A182N810 A0A182LUC3 A0A182RTP1 A0A131Y645 A0A0B6YA19 B7PEL3 A0A1Q3FBD0 Q5ZK10 A0A1L8EII4 A0A1A9X3S0 D3TRN7 A0A1L8EIS2 A0A3M0JA46 B5FYZ6 B5KFR1 A0A1L8EIN0 A0A1A7XN86 A0A226NJI6 H0Z9A8 A0A3L8SAB3 Q7Q9P4 A0A218UT36 B0WHN1 A0A1B0A9G6 J3JUH8 A0A182HP69 A0A182WAA6 A0A2I4BBC2 A0A1A9USQ5 A0A3S1ABH4
A0A2W1BD25 A0A1B6KXR7 A0A2J7PD98 A0A2J7PDA4 A0A1Y1L4L0 D6WT40 A0A151IV27 A0A3L8DFP9 A0A195F004 E2AKL7 F4WAT6 A0A151XDM5 A0A195BWY2 A0A151IH76 A0A158P3K4 E9J8R6 A0A182GLM9 Q1HRG9 A0A0J7LAA6 A0A087ZQ92 A0A162D7N3 A0A232FC07 A0A1W4WTH1 A0A2A3EJ17 V9IDV9 A0A310S969 E2BNT9 K7J497 A0A0M9A2E6 E9FYH5 U5EYU9 A0A0K8TSI9 A0A0P4W4J8 A0A0P5CXR9 A0A182FH85 A0A154PIP1 V5H138 A0A1I8M796 T1PDZ5 A0A2Y9F8U5 A0A2Y9PSG9 A0A341B7H5 A0A2U3V8Q1 A0A084W9K5 A0A2M4BQM0 A0A2M4ADL1 A0A067R7M4 A0A2M4BQE1 A0A2Y9PTH9 A0A0L7R492 A0A2Y9F790 A0A341BAU6 A0A2U3V8B0 A0A1S2ZQU1 A0A2U4CLV0 A0A341BAU0 A0A2Y9PYU9 A0A340Y1D4 C3Y288 W5JP15 A0A3Q2XMI0 A0A182N810 A0A182LUC3 A0A182RTP1 A0A131Y645 A0A0B6YA19 B7PEL3 A0A1Q3FBD0 Q5ZK10 A0A1L8EII4 A0A1A9X3S0 D3TRN7 A0A1L8EIS2 A0A3M0JA46 B5FYZ6 B5KFR1 A0A1L8EIN0 A0A1A7XN86 A0A226NJI6 H0Z9A8 A0A3L8SAB3 Q7Q9P4 A0A218UT36 B0WHN1 A0A1B0A9G6 J3JUH8 A0A182HP69 A0A182WAA6 A0A2I4BBC2 A0A1A9USQ5 A0A3S1ABH4
Pubmed
EMBL
NWSH01003367
PCG66447.1
RSAL01000029
RVE51751.1
GAIX01003953
JAA88607.1
+ More
ODYU01002757 SOQ40647.1 PCG66446.1 GDQN01007215 JAT83839.1 KZ150264 PZC71714.1 GEBQ01023739 JAT16238.1 NEVH01026392 PNF14316.1 PNF14314.1 GEZM01064873 GEZM01064872 JAV68623.1 KQ971354 EFA05869.1 KQ980924 KYN11414.1 QOIP01000008 RLU19224.1 KQ981880 KYN33900.1 GL440320 EFN66021.1 GL888053 EGI68692.1 KQ982268 KYQ58465.1 KQ976394 KYM93139.1 KQ977636 KYN01145.1 ADTU01008167 GL769036 EFZ10817.1 JXUM01072366 KQ562717 KXJ75261.1 DQ440125 ABF18158.1 LBMM01000143 KMR04773.1 LRGB01002371 KZS08034.1 NNAY01000441 OXU28374.1 KZ288229 PBC31717.1 JR039309 AEY58631.1 KQ767768 OAD53308.1 GL449507 EFN82622.1 AAZX01007448 KQ435756 KOX75895.1 GL732527 EFX87766.1 GANO01000061 JAB59810.1 GDAI01000266 JAI17337.1 GDRN01085026 JAI61413.1 GDIP01163948 JAJ59454.1 KQ434904 KZC11188.1 GALX01001913 JAB66553.1 KA646924 AFP61553.1 ATLV01021829 KE525324 KFB46899.1 GGFJ01006163 MBW55304.1 GGFK01005553 MBW38874.1 KK852647 KDR19517.1 GGFJ01006138 MBW55279.1 KQ414658 KOC65644.1 GG666480 EEN65978.1 ADMH02000991 ETN64499.1 AXCM01000363 GEFM01001946 JAP73850.1 HACG01005435 CEK52300.1 ABJB010223645 DS695957 EEC05035.1 GFDL01010242 JAV24803.1 AJ720274 GFDG01000282 JAV18517.1 EZ424089 ADD20365.1 GFDG01000280 JAV18519.1 QRBI01000184 RMB95763.1 DQ214244 ACH44257.1 EF191934 ACH46409.1 GFDG01000281 JAV18518.1 HADW01018152 HADX01010320 SBP19552.1 MCFN01000030 OXB67833.1 ABQF01028308 QUSF01000037 RLV98843.1 AAAB01008900 EAA09447.4 MUZQ01000152 OWK56590.1 DS231938 EDS27880.1 BT126892 AEE61854.1 APCN01002922 RQTK01000108 RUS87533.1
ODYU01002757 SOQ40647.1 PCG66446.1 GDQN01007215 JAT83839.1 KZ150264 PZC71714.1 GEBQ01023739 JAT16238.1 NEVH01026392 PNF14316.1 PNF14314.1 GEZM01064873 GEZM01064872 JAV68623.1 KQ971354 EFA05869.1 KQ980924 KYN11414.1 QOIP01000008 RLU19224.1 KQ981880 KYN33900.1 GL440320 EFN66021.1 GL888053 EGI68692.1 KQ982268 KYQ58465.1 KQ976394 KYM93139.1 KQ977636 KYN01145.1 ADTU01008167 GL769036 EFZ10817.1 JXUM01072366 KQ562717 KXJ75261.1 DQ440125 ABF18158.1 LBMM01000143 KMR04773.1 LRGB01002371 KZS08034.1 NNAY01000441 OXU28374.1 KZ288229 PBC31717.1 JR039309 AEY58631.1 KQ767768 OAD53308.1 GL449507 EFN82622.1 AAZX01007448 KQ435756 KOX75895.1 GL732527 EFX87766.1 GANO01000061 JAB59810.1 GDAI01000266 JAI17337.1 GDRN01085026 JAI61413.1 GDIP01163948 JAJ59454.1 KQ434904 KZC11188.1 GALX01001913 JAB66553.1 KA646924 AFP61553.1 ATLV01021829 KE525324 KFB46899.1 GGFJ01006163 MBW55304.1 GGFK01005553 MBW38874.1 KK852647 KDR19517.1 GGFJ01006138 MBW55279.1 KQ414658 KOC65644.1 GG666480 EEN65978.1 ADMH02000991 ETN64499.1 AXCM01000363 GEFM01001946 JAP73850.1 HACG01005435 CEK52300.1 ABJB010223645 DS695957 EEC05035.1 GFDL01010242 JAV24803.1 AJ720274 GFDG01000282 JAV18517.1 EZ424089 ADD20365.1 GFDG01000280 JAV18519.1 QRBI01000184 RMB95763.1 DQ214244 ACH44257.1 EF191934 ACH46409.1 GFDG01000281 JAV18518.1 HADW01018152 HADX01010320 SBP19552.1 MCFN01000030 OXB67833.1 ABQF01028308 QUSF01000037 RLV98843.1 AAAB01008900 EAA09447.4 MUZQ01000152 OWK56590.1 DS231938 EDS27880.1 BT126892 AEE61854.1 APCN01002922 RQTK01000108 RUS87533.1
Proteomes
UP000218220
UP000283053
UP000235965
UP000007266
UP000078492
UP000279307
+ More
UP000078541 UP000000311 UP000007755 UP000075809 UP000078540 UP000078542 UP000005205 UP000069940 UP000249989 UP000036403 UP000005203 UP000076858 UP000215335 UP000192223 UP000242457 UP000008237 UP000002358 UP000053105 UP000000305 UP000069272 UP000076502 UP000095301 UP000248484 UP000248483 UP000252040 UP000245320 UP000030765 UP000027135 UP000053825 UP000079721 UP000265300 UP000001554 UP000000673 UP000264820 UP000075884 UP000075883 UP000075900 UP000001555 UP000000539 UP000091820 UP000269221 UP000198323 UP000007754 UP000276834 UP000007062 UP000197619 UP000002320 UP000092445 UP000075840 UP000075920 UP000192220 UP000078200 UP000271974
UP000078541 UP000000311 UP000007755 UP000075809 UP000078540 UP000078542 UP000005205 UP000069940 UP000249989 UP000036403 UP000005203 UP000076858 UP000215335 UP000192223 UP000242457 UP000008237 UP000002358 UP000053105 UP000000305 UP000069272 UP000076502 UP000095301 UP000248484 UP000248483 UP000252040 UP000245320 UP000030765 UP000027135 UP000053825 UP000079721 UP000265300 UP000001554 UP000000673 UP000264820 UP000075884 UP000075883 UP000075900 UP000001555 UP000000539 UP000091820 UP000269221 UP000198323 UP000007754 UP000276834 UP000007062 UP000197619 UP000002320 UP000092445 UP000075840 UP000075920 UP000192220 UP000078200 UP000271974
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4J2R8
A0A3S2P3Y6
S4PY40
A0A2H1VIJ3
A0A2A4J3Y5
A0A1E1WA73
+ More
A0A2W1BD25 A0A1B6KXR7 A0A2J7PD98 A0A2J7PDA4 A0A1Y1L4L0 D6WT40 A0A151IV27 A0A3L8DFP9 A0A195F004 E2AKL7 F4WAT6 A0A151XDM5 A0A195BWY2 A0A151IH76 A0A158P3K4 E9J8R6 A0A182GLM9 Q1HRG9 A0A0J7LAA6 A0A087ZQ92 A0A162D7N3 A0A232FC07 A0A1W4WTH1 A0A2A3EJ17 V9IDV9 A0A310S969 E2BNT9 K7J497 A0A0M9A2E6 E9FYH5 U5EYU9 A0A0K8TSI9 A0A0P4W4J8 A0A0P5CXR9 A0A182FH85 A0A154PIP1 V5H138 A0A1I8M796 T1PDZ5 A0A2Y9F8U5 A0A2Y9PSG9 A0A341B7H5 A0A2U3V8Q1 A0A084W9K5 A0A2M4BQM0 A0A2M4ADL1 A0A067R7M4 A0A2M4BQE1 A0A2Y9PTH9 A0A0L7R492 A0A2Y9F790 A0A341BAU6 A0A2U3V8B0 A0A1S2ZQU1 A0A2U4CLV0 A0A341BAU0 A0A2Y9PYU9 A0A340Y1D4 C3Y288 W5JP15 A0A3Q2XMI0 A0A182N810 A0A182LUC3 A0A182RTP1 A0A131Y645 A0A0B6YA19 B7PEL3 A0A1Q3FBD0 Q5ZK10 A0A1L8EII4 A0A1A9X3S0 D3TRN7 A0A1L8EIS2 A0A3M0JA46 B5FYZ6 B5KFR1 A0A1L8EIN0 A0A1A7XN86 A0A226NJI6 H0Z9A8 A0A3L8SAB3 Q7Q9P4 A0A218UT36 B0WHN1 A0A1B0A9G6 J3JUH8 A0A182HP69 A0A182WAA6 A0A2I4BBC2 A0A1A9USQ5 A0A3S1ABH4
A0A2W1BD25 A0A1B6KXR7 A0A2J7PD98 A0A2J7PDA4 A0A1Y1L4L0 D6WT40 A0A151IV27 A0A3L8DFP9 A0A195F004 E2AKL7 F4WAT6 A0A151XDM5 A0A195BWY2 A0A151IH76 A0A158P3K4 E9J8R6 A0A182GLM9 Q1HRG9 A0A0J7LAA6 A0A087ZQ92 A0A162D7N3 A0A232FC07 A0A1W4WTH1 A0A2A3EJ17 V9IDV9 A0A310S969 E2BNT9 K7J497 A0A0M9A2E6 E9FYH5 U5EYU9 A0A0K8TSI9 A0A0P4W4J8 A0A0P5CXR9 A0A182FH85 A0A154PIP1 V5H138 A0A1I8M796 T1PDZ5 A0A2Y9F8U5 A0A2Y9PSG9 A0A341B7H5 A0A2U3V8Q1 A0A084W9K5 A0A2M4BQM0 A0A2M4ADL1 A0A067R7M4 A0A2M4BQE1 A0A2Y9PTH9 A0A0L7R492 A0A2Y9F790 A0A341BAU6 A0A2U3V8B0 A0A1S2ZQU1 A0A2U4CLV0 A0A341BAU0 A0A2Y9PYU9 A0A340Y1D4 C3Y288 W5JP15 A0A3Q2XMI0 A0A182N810 A0A182LUC3 A0A182RTP1 A0A131Y645 A0A0B6YA19 B7PEL3 A0A1Q3FBD0 Q5ZK10 A0A1L8EII4 A0A1A9X3S0 D3TRN7 A0A1L8EIS2 A0A3M0JA46 B5FYZ6 B5KFR1 A0A1L8EIN0 A0A1A7XN86 A0A226NJI6 H0Z9A8 A0A3L8SAB3 Q7Q9P4 A0A218UT36 B0WHN1 A0A1B0A9G6 J3JUH8 A0A182HP69 A0A182WAA6 A0A2I4BBC2 A0A1A9USQ5 A0A3S1ABH4
PDB
1S3S
E-value=9.79086e-19,
Score=229
Ontologies
GO
Topology
Subcellular location
Nucleus
Predominantly nuclear in interphase cells. A small proportion of the protein is cytoplasmic, associated with Golgi stacks. Localizes to centrosome during mitotic prophase and metaphase. With evidence from 1 publications.
Golgi apparatus Predominantly nuclear in interphase cells. A small proportion of the protein is cytoplasmic, associated with Golgi stacks. Localizes to centrosome during mitotic prophase and metaphase. With evidence from 1 publications.
Golgi stack Predominantly nuclear in interphase cells. A small proportion of the protein is cytoplasmic, associated with Golgi stacks. Localizes to centrosome during mitotic prophase and metaphase. With evidence from 1 publications.
Cytoplasm Predominantly nuclear in interphase cells. A small proportion of the protein is cytoplasmic, associated with Golgi stacks. Localizes to centrosome during mitotic prophase and metaphase. With evidence from 1 publications.
Cytoskeleton Predominantly nuclear in interphase cells. A small proportion of the protein is cytoplasmic, associated with Golgi stacks. Localizes to centrosome during mitotic prophase and metaphase. With evidence from 1 publications.
Microtubule organizing center Predominantly nuclear in interphase cells. A small proportion of the protein is cytoplasmic, associated with Golgi stacks. Localizes to centrosome during mitotic prophase and metaphase. With evidence from 1 publications.
Centrosome Predominantly nuclear in interphase cells. A small proportion of the protein is cytoplasmic, associated with Golgi stacks. Localizes to centrosome during mitotic prophase and metaphase. With evidence from 1 publications.
Golgi apparatus Predominantly nuclear in interphase cells. A small proportion of the protein is cytoplasmic, associated with Golgi stacks. Localizes to centrosome during mitotic prophase and metaphase. With evidence from 1 publications.
Golgi stack Predominantly nuclear in interphase cells. A small proportion of the protein is cytoplasmic, associated with Golgi stacks. Localizes to centrosome during mitotic prophase and metaphase. With evidence from 1 publications.
Cytoplasm Predominantly nuclear in interphase cells. A small proportion of the protein is cytoplasmic, associated with Golgi stacks. Localizes to centrosome during mitotic prophase and metaphase. With evidence from 1 publications.
Cytoskeleton Predominantly nuclear in interphase cells. A small proportion of the protein is cytoplasmic, associated with Golgi stacks. Localizes to centrosome during mitotic prophase and metaphase. With evidence from 1 publications.
Microtubule organizing center Predominantly nuclear in interphase cells. A small proportion of the protein is cytoplasmic, associated with Golgi stacks. Localizes to centrosome during mitotic prophase and metaphase. With evidence from 1 publications.
Centrosome Predominantly nuclear in interphase cells. A small proportion of the protein is cytoplasmic, associated with Golgi stacks. Localizes to centrosome during mitotic prophase and metaphase. With evidence from 1 publications.
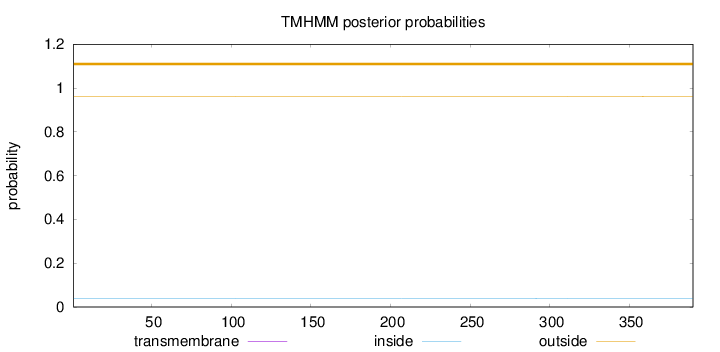
Length:
390
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00237
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03915
outside
1 - 390
Population Genetic Test Statistics
Pi
229.446622
Theta
192.973395
Tajima's D
1.620957
CLR
0.544854
CSRT
0.814609269536523
Interpretation
Uncertain
