Gene
KWMTBOMO16308
Pre Gene Modal
BGIBMGA013768
Annotation
PREDICTED:_uncharacterized_protein_LOC101738379_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.988
Sequence
CDS
ATGAAGAATCAGCGCAAATCATCCACAGGAATCATGAGGAAATATTACCAACTTATTCTCATCGTCATCTGTCTGATCAGCGTAGTAACGTTACTCATATACCGCCACGAATACTACCGGCTTCGTTATGTATTGGAAGTGCTTAATTTCTTTGGAAAACCTGGACTGTCGGAGATCGAATTTTGCGGACCCGGTTTCAACGCTACAATGCTATCAGAAATACTTAAGAACTCATCCATGGAGATAAGAGAAACGCCACCCTTGTTTCAACAAATAGATGAAAACTTTTATGCATACACATCATTCTTGAAGTCATATCAAAAATATAACAAGTTGACCCCATCCGATGTACACAATATAGATACGATAGTAATCGGTAAGGCGCAGAGTGAACCCAGTTTCCGCTGTAATATTTGGTTTGACAACAATAATAAACCCAAAGCAGGAAAATTCAGTTATAGAGCTATCACAGAACCAGTTGGTGAGTTCAGAGTTTATGTCTTCCAATGTTCACTTAACAAGAATTTAGGTAAACCTAAAGGAGTAAGTTTTTATGTAAATGATTATAATATTAATCCCATACATGCACCGATTAATAGACTTATTGCGGTTAATACAAAAAGGAAAGTTAGGGAAAGTAGTAGTGTAAAGTTTGTCAATAATATTGTACCAGCAATTTGTGTTATACCCAACCAAGTACCGATGGTCACGAGGGACAATTTTATAGAATTTTTAGTTTTCCATCACATGATGGGATTTGATTATTTCACTATTTATGATAGTATGATCTCGGAGGATATAATTAGGAGACTGAACTTAATACCTTCAGATATAACTCAGTGGAAAATACAATTTTTCCCTCTGAATTTTCCGTTTGTTTTTGCAAAATCATATGAAGTAGTACGAAGTGCTATAGAATTAGATTGCTTGTTCAGACACTTTAGACTAGACAAAGAAGATTTAGAGAAAGTTAGCCATGTCGTAACTTTATCTTGGGACGAGTTTTTGGTTCCTAGAGTTTATAGTAATGTGAAGGCAATTATTGACGACCTAGACCCAACCAGAAAAGTACACAGTTTTGAAGTTAAACCATTACTGTTCTGTCTGAATCAAAATGACGATGATAACACAAATATTGCCTATCCAGATATAATGAAGAAAACACACTACTATATAATTCCAAATCAACAGATAAAATCTCTGCATATGCGAAATTTGGATGCGATGAAAACGTTTGATGATATTTTAAATTTGAATTCTAGTAGAATAATACCGTTGGAGACATTAGGTGCCCATAAGTATACTGAATGTAGGGATCCTAAAACGTACCAACCTGAATCCTACAATGAGGAACAGATGCAAGGCTTTAAACACAGAATGGAGGGAGCTATGCTTAGTTTTGCTGAAAGTTTAAATAGTAACAAGATGTACAGACTGTATAGAGCTGGACAAATATGGGAAAAGACACCCAGTGACGAAAAAGATTTGTAA
Protein
MKNQRKSSTGIMRKYYQLILIVICLISVVTLLIYRHEYYRLRYVLEVLNFFGKPGLSEIEFCGPGFNATMLSEILKNSSMEIRETPPLFQQIDENFYAYTSFLKSYQKYNKLTPSDVHNIDTIVIGKAQSEPSFRCNIWFDNNNKPKAGKFSYRAITEPVGEFRVYVFQCSLNKNLGKPKGVSFYVNDYNINPIHAPINRLIAVNTKRKVRESSSVKFVNNIVPAICVIPNQVPMVTRDNFIEFLVFHHMMGFDYFTIYDSMISEDIIRRLNLIPSDITQWKIQFFPLNFPFVFAKSYEVVRSAIELDCLFRHFRLDKEDLEKVSHVVTLSWDEFLVPRVYSNVKAIIDDLDPTRKVHSFEVKPLLFCLNQNDDDNTNIAYPDIMKKTHYYIIPNQQIKSLHMRNLDAMKTFDDILNLNSSRIIPLETLGAHKYTECRDPKTYQPESYNEEQMQGFKHRMEGAMLSFAESLNSNKMYRLYRAGQIWEKTPSDEKDL
Summary
Uniprot
A0A2W1BJ76
A0A2A4J440
A0A0L7L5E6
A0A3S2NMY2
A0A2H1VIM6
A0A0N1IES2
+ More
A0A0N1PE78 A0A2P8Z396 A0A067QII3 A0A232FCZ4 A0A2J7PD94 A0A154PHA5 K7J498 E2AKL8 A0A195F1B9 A0A158P3K3 A0A087ZQ63 A0A151IV53 A0A0L7R470 E9J8R7 A0A151IH61 A0A2A3EKK3 A0A0N0U5T8 A0A1B6CFK3 E0VQG5 A0A026X0H5 A0A310SGI3 E2BNT8 A0A1B0GNQ3 A0A1B0BE80 D6WQD5 A0A1B6HBB2 A0A1B0CXA0 A0A1A9USQ9 A0A1B6M3E2 B4ME28 B4J7W0 A0A0V0G887 A0A1L8DCB1 A0A1B0G5V0 D3TQI2 A0A224XDL7 A0A1I8P171 A0A0P4VX03 A0A1W4WAH4 R4FK84 W8BSX1 B4KQI7 A0A3Q0JIJ2 A0A1I8MDG4 A0A0L0C826 A0A0C9R1Y1
A0A0N1PE78 A0A2P8Z396 A0A067QII3 A0A232FCZ4 A0A2J7PD94 A0A154PHA5 K7J498 E2AKL8 A0A195F1B9 A0A158P3K3 A0A087ZQ63 A0A151IV53 A0A0L7R470 E9J8R7 A0A151IH61 A0A2A3EKK3 A0A0N0U5T8 A0A1B6CFK3 E0VQG5 A0A026X0H5 A0A310SGI3 E2BNT8 A0A1B0GNQ3 A0A1B0BE80 D6WQD5 A0A1B6HBB2 A0A1B0CXA0 A0A1A9USQ9 A0A1B6M3E2 B4ME28 B4J7W0 A0A0V0G887 A0A1L8DCB1 A0A1B0G5V0 D3TQI2 A0A224XDL7 A0A1I8P171 A0A0P4VX03 A0A1W4WAH4 R4FK84 W8BSX1 B4KQI7 A0A3Q0JIJ2 A0A1I8MDG4 A0A0L0C826 A0A0C9R1Y1
Pubmed
EMBL
KZ150264
PZC71713.1
NWSH01003367
PCG66448.1
JTDY01002814
KOB70667.1
+ More
RSAL01000029 RVE51750.1 ODYU01002757 SOQ40646.1 KQ460460 KPJ14632.1 KQ459166 KPJ03580.1 PYGN01000217 PSN50974.1 KK853536 KDR06922.1 NNAY01000441 OXU28370.1 NEVH01026392 PNF14305.1 KQ434904 KZC11187.1 AAZX01006624 AAZX01007448 GL440320 EFN66022.1 KQ981880 KYN33899.1 ADTU01008167 KQ980924 KYN11413.1 KQ414658 KOC65643.1 GL769036 EFZ10804.1 KQ977636 KYN01144.1 KZ288229 PBC31716.1 KQ435756 KOX75894.1 GEDC01025086 JAS12212.1 DS235430 EEB15621.1 KK107064 QOIP01000008 EZA60899.1 RLU19491.1 KQ767768 OAD53309.1 GL449507 EFN82621.1 AJVK01029673 JXJN01012806 KQ971354 EFA06088.1 GECU01035728 JAS71978.1 AJWK01033471 GEBQ01009567 JAT30410.1 CH940662 EDW58793.1 CH916367 EDW01166.1 GECL01001894 JAP04230.1 GFDF01010124 JAV03960.1 CCAG010007475 EZ423684 ADD19960.1 GFTR01005881 JAW10545.1 GDKW01001385 JAI55210.1 ACPB03011931 GAHY01001973 JAA75537.1 GAMC01014241 JAB92314.1 CH933808 EDW08156.1 JRES01000780 KNC28382.1 GBYB01006830 GBYB01006832 JAG76597.1 JAG76599.1
RSAL01000029 RVE51750.1 ODYU01002757 SOQ40646.1 KQ460460 KPJ14632.1 KQ459166 KPJ03580.1 PYGN01000217 PSN50974.1 KK853536 KDR06922.1 NNAY01000441 OXU28370.1 NEVH01026392 PNF14305.1 KQ434904 KZC11187.1 AAZX01006624 AAZX01007448 GL440320 EFN66022.1 KQ981880 KYN33899.1 ADTU01008167 KQ980924 KYN11413.1 KQ414658 KOC65643.1 GL769036 EFZ10804.1 KQ977636 KYN01144.1 KZ288229 PBC31716.1 KQ435756 KOX75894.1 GEDC01025086 JAS12212.1 DS235430 EEB15621.1 KK107064 QOIP01000008 EZA60899.1 RLU19491.1 KQ767768 OAD53309.1 GL449507 EFN82621.1 AJVK01029673 JXJN01012806 KQ971354 EFA06088.1 GECU01035728 JAS71978.1 AJWK01033471 GEBQ01009567 JAT30410.1 CH940662 EDW58793.1 CH916367 EDW01166.1 GECL01001894 JAP04230.1 GFDF01010124 JAV03960.1 CCAG010007475 EZ423684 ADD19960.1 GFTR01005881 JAW10545.1 GDKW01001385 JAI55210.1 ACPB03011931 GAHY01001973 JAA75537.1 GAMC01014241 JAB92314.1 CH933808 EDW08156.1 JRES01000780 KNC28382.1 GBYB01006830 GBYB01006832 JAG76597.1 JAG76599.1
Proteomes
UP000218220
UP000037510
UP000283053
UP000053240
UP000053268
UP000245037
+ More
UP000027135 UP000215335 UP000235965 UP000076502 UP000002358 UP000000311 UP000078541 UP000005205 UP000005203 UP000078492 UP000053825 UP000078542 UP000242457 UP000053105 UP000009046 UP000053097 UP000279307 UP000008237 UP000092462 UP000092460 UP000007266 UP000092461 UP000078200 UP000008792 UP000001070 UP000092444 UP000095300 UP000192223 UP000015103 UP000009192 UP000079169 UP000095301 UP000037069
UP000027135 UP000215335 UP000235965 UP000076502 UP000002358 UP000000311 UP000078541 UP000005205 UP000005203 UP000078492 UP000053825 UP000078542 UP000242457 UP000053105 UP000009046 UP000053097 UP000279307 UP000008237 UP000092462 UP000092460 UP000007266 UP000092461 UP000078200 UP000008792 UP000001070 UP000092444 UP000095300 UP000192223 UP000015103 UP000009192 UP000079169 UP000095301 UP000037069
Interpro
SUPFAM
SSF100879
SSF100879
Gene 3D
ProteinModelPortal
A0A2W1BJ76
A0A2A4J440
A0A0L7L5E6
A0A3S2NMY2
A0A2H1VIM6
A0A0N1IES2
+ More
A0A0N1PE78 A0A2P8Z396 A0A067QII3 A0A232FCZ4 A0A2J7PD94 A0A154PHA5 K7J498 E2AKL8 A0A195F1B9 A0A158P3K3 A0A087ZQ63 A0A151IV53 A0A0L7R470 E9J8R7 A0A151IH61 A0A2A3EKK3 A0A0N0U5T8 A0A1B6CFK3 E0VQG5 A0A026X0H5 A0A310SGI3 E2BNT8 A0A1B0GNQ3 A0A1B0BE80 D6WQD5 A0A1B6HBB2 A0A1B0CXA0 A0A1A9USQ9 A0A1B6M3E2 B4ME28 B4J7W0 A0A0V0G887 A0A1L8DCB1 A0A1B0G5V0 D3TQI2 A0A224XDL7 A0A1I8P171 A0A0P4VX03 A0A1W4WAH4 R4FK84 W8BSX1 B4KQI7 A0A3Q0JIJ2 A0A1I8MDG4 A0A0L0C826 A0A0C9R1Y1
A0A0N1PE78 A0A2P8Z396 A0A067QII3 A0A232FCZ4 A0A2J7PD94 A0A154PHA5 K7J498 E2AKL8 A0A195F1B9 A0A158P3K3 A0A087ZQ63 A0A151IV53 A0A0L7R470 E9J8R7 A0A151IH61 A0A2A3EKK3 A0A0N0U5T8 A0A1B6CFK3 E0VQG5 A0A026X0H5 A0A310SGI3 E2BNT8 A0A1B0GNQ3 A0A1B0BE80 D6WQD5 A0A1B6HBB2 A0A1B0CXA0 A0A1A9USQ9 A0A1B6M3E2 B4ME28 B4J7W0 A0A0V0G887 A0A1L8DCB1 A0A1B0G5V0 D3TQI2 A0A224XDL7 A0A1I8P171 A0A0P4VX03 A0A1W4WAH4 R4FK84 W8BSX1 B4KQI7 A0A3Q0JIJ2 A0A1I8MDG4 A0A0L0C826 A0A0C9R1Y1
Ontologies
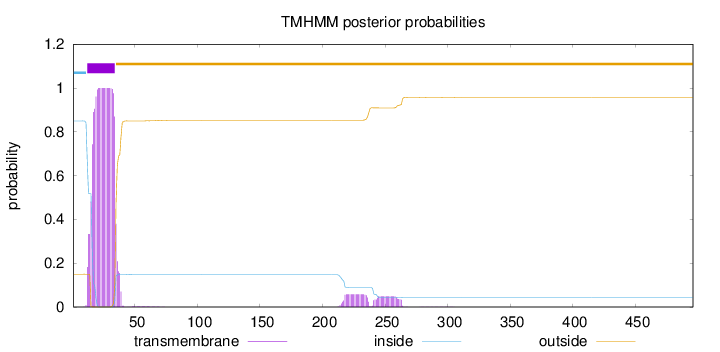
Topology
Length:
496
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.9372
Exp number, first 60 AAs:
20.69846
Total prob of N-in:
0.84987
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 34
outside
35 - 496
Population Genetic Test Statistics
Pi
171.213956
Theta
204.031679
Tajima's D
-0.620597
CLR
806.208563
CSRT
0.214489275536223
Interpretation
Uncertain
