Gene
KWMTBOMO16306
Pre Gene Modal
BGIBMGA013771
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_A-kinase_anchor_protein_13_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.57
Sequence
CDS
ATGAACTTATATTTAAATAAAAAAAGCTTGTCTGGCGCCGGTGAGAGCGAGACGGAAAGCGACGAAGACGCTTCCGCCGCGACTGTCGGGGCCACGGCACAACTGCCCGGCCGCAGGAACAATACGCATCAGGAACATTTGGGATTACATCGACTGGCTTTCTTGGAACAAGCTGCTTTTGACCATCACGCTGAGAAAAGGCGAAAACGTGGAAGCTTGTTCTTCAGGAAGAAGAAGGACAAGTCCCGGAAGGCGGCGCACGCGTGGCAGGCGGGCGGCGGCGCGCACTGCGACTGGTGCTCGCGACCCCTCAACGACACGCCCGCCCTCTACTGCGACAATTGTGGAACAACTGTACATCAAAATATTTGCAAAGATTACATCGTCGATTGCAATAAGCCTAAGTCGTCTAAGACATCTGTTGGTAAATCACTGAGTGCAGCCAGCGGGAAAAGCAGCAAACGGAGCTCCGTATCCGGACAGTCGCAGACTAATAGCACCAGTCATATTTATAACGACGACAAGGATTCAGATCATAAACACGACCAGAGCAACGCTTCGGACGACGCGTGCGCCGCGGAGTGGCCCGAGGTGTATCTGGGCCCCGAGCAGCTGGGCGACGAGGCGCTGGTGCTGGGGCTGGGCGCCAGCGAGCCCGACACCTGGGCGGCCGGCGCTCCCAAGGGACTCGCCAAGTCGCTCGGCGAAAGAGAGACGAAGCGACAAGAACACATTTACGAGCTGATACTCACGGAGAAGCATCACTGTCTCACCATACGACTTATGCAAAAGATGTTTGCTGACGGCATGGTGCGGTTCGGCAGCGTGAGCTCGGCGCAGGTGGCGCGCATGTTCCCGCGCGTGGACGAGCTGTGGGCGCTGCACGCGGCGCTGCTGGCCCGCCTGCGCGCCCGCCAGCGCGCCGCGCCCGCCGTGCCCTCCATCGCCGACATCCTGGCCGACGCCTTCGCGCCGCACTCCCGCCCGCGCCTCAAGTCCGCCTACGGCGAGTTTTGCTCCCGTCACCGCGACGCCGTGGAGGTGTTTAAGGAGTGCTGCGCGAGAGACGCCCGCCTGTCCCGGTTCATCAGGAAGTGCCAACAGAACCCTTTGCTCAGGAAGAAGGGTGTTCCCGAGTGCGTGCTGTTCGTAGCGCAGCGACTCACCAAGTACCCGCTGCTGCTGGAGCCTCTACTCAAGACGGCCGGCGATGACGTCAACGAACGTGAGCTGCTGCAGAAGGCGCTCTGCGGAGTCAAGGAGATCCTAGTGGATGTAGACAACCAGGTGGCGGCGAAGGAGCGCGAGGACCGCAAGCTCGAGATCTACCACCGCATCGACGCCAAGTCCTTCGCTCACTTCCGCGGCAGGAAGTTCAAGAAGAGCGACATACTGCAGGGCACCAGGAGCTTAAAGTTTGAAGGTGTGGCCACACTGATGCAAGGCCGCAGCAAGATGCAAACCCTACTGGTGATCGTGCTCTCCGACGTGCTGTTCTTCCTGCACGACAACAACAATAAATACACGTTCTTCACGCCCGACAATAAAACCGGTGTCGTGTCTCTCGTGAAGCTACTGGTCCGCGAGAAGGCGGGTGCTGAAGGCAGGGGCTTGTATCTGATCTGCAGCGGCCCCAACGAACCAGAAATGTTCGAGCTGCGAGTTCACAGGCCTAAGGATATACAGACTTGGATACAGGCCATAAGAGCCGCAGTCCAGAACTGCCCTGAGGAGGTGGAAGAGACCGAGGCAGGGCTGTTGGCTACCTCCGCGGAGGAGAGACAACGCCAGCTGGAAGCCAGACACGAGAATATAAGACTGATCACAGAAGCGTTAAGGACAAAAGACAAGGAGCTAGCGACACTCCTTGAAGAAAAATTGGCGCTGCACATGAAGATGGTGGGCCACACCGGAACCTCTACGCTGGACGTGCCTCACACGGGACTAAACACGTCATCGCCCGGGGGCTTGGTGTTTCCAGACTACGTGAGGCTGGTGGCGCCATCTATAGATTCGCATACAATATGGCAGGAAGTTTGTCGCGTTGTCAAGGAGGCGCTGGAGAGTGCTGGGTGCGCGTGGGGCGGCGCCCTGGGCCGCAGCACCAGCTCGGCGGGCGAGCGGCACTCCGCCGCCTACACCAGCCCCGCGCTGCCGCGCCGCGCCGACACCTTCGCGGGGTTCGACGCCACCAAGCACAGGGGTCTCGTGCACTGCTCTCGATAA
Protein
MNLYLNKKSLSGAGESETESDEDASAATVGATAQLPGRRNNTHQEHLGLHRLAFLEQAAFDHHAEKRRKRGSLFFRKKKDKSRKAAHAWQAGGGAHCDWCSRPLNDTPALYCDNCGTTVHQNICKDYIVDCNKPKSSKTSVGKSLSAASGKSSKRSSVSGQSQTNSTSHIYNDDKDSDHKHDQSNASDDACAAEWPEVYLGPEQLGDEALVLGLGASEPDTWAAGAPKGLAKSLGERETKRQEHIYELILTEKHHCLTIRLMQKMFADGMVRFGSVSSAQVARMFPRVDELWALHAALLARLRARQRAAPAVPSIADILADAFAPHSRPRLKSAYGEFCSRHRDAVEVFKECCARDARLSRFIRKCQQNPLLRKKGVPECVLFVAQRLTKYPLLLEPLLKTAGDDVNERELLQKALCGVKEILVDVDNQVAAKEREDRKLEIYHRIDAKSFAHFRGRKFKKSDILQGTRSLKFEGVATLMQGRSKMQTLLVIVLSDVLFFLHDNNNKYTFFTPDNKTGVVSLVKLLVREKAGAEGRGLYLICSGPNEPEMFELRVHRPKDIQTWIQAIRAAVQNCPEEVEETEAGLLATSAEERQRQLEARHENIRLITEALRTKDKELATLLEEKLALHMKMVGHTGTSTLDVPHTGLNTSSPGGLVFPDYVRLVAPSIDSHTIWQEVCRVVKEALESAGCAWGGALGRSTSSAGERHSAAYTSPALPRRADTFAGFDATKHRGLVHCSR
Summary
Uniprot
A0A3S2LE36
A0A0N1IA73
H9JW58
A0A2A4J4L6
A0A2A4J4T9
A0A2A4J4X7
+ More
A0A2A4J373 A0A2A4J3K4 A0A2A4J4W0 A0A2W1BG02 A0A1Y1N5S4 A0A1Y1N8Q1 A0A1Y1NAJ4 A0A1Y1N5N1 A0A1Y1N680 A0A1Y1NC98 A0A1Y1N719 A0A1Y1N9E0 A0A2J7R6J0 A0A2J7R6K3 A0A2J7R6J4 A0A2J7R6J7 D6WQX0 E0VAR1 A0A067QPR7 A0A0A9WVI4 A0A1Y1N5V1 N6SW71 A0A2S2PPT7 A0A2S2QXJ5 J9K9G0 A0A2J7R6J8 A0A2H8TN70 A0A2J7R6K8 E2B806 A0A154P8A3 A0A224XDS9 F4WP99 A0A195E5P1 A0A336LX67 A0A151WMJ0 A0A195EUJ2 A0A336LKL2 A0A336LPD6 A0A336LJW4 A0A336K0D9 A0A336K2W3 A0A026WNT8 A0A195C8C2 A0A0L7R928 A0A195B125 A0A0N8ES16 A0A1S4FZ82 Q16J05 A0A0C9QND1 A0A087ZX61 A0A310SCC2 B0WVU5 A0A0C9R5J4 K7IZC0 A0A1Q3EXQ7 A0A2M4BAJ0 A0A1Q3EXS2 A0A232EX52 A0A158NR65 A0A1Q3EXL2 A0A182FG24 A0A2M4A7J3 A0A2M4B994 A0A2M4B9A7 A0A2M4B961 W5JH27 E9J571 A0A182T4A1 A0A3L8DWX1 A0A182NHB7 A0A182QXC4 Q7QJX4 A0A1L8DLM0 E2ADU0 A0A084VF60 A0A182RB14 A0A182WIZ1 A0A182LWP7 V9IKV3 A0A0J7NI72 A0A182IM63 A0A0C9PM36 A0A182JQW3 A0A0A9VU05 A0A146MC82 W4VRK5
A0A2A4J373 A0A2A4J3K4 A0A2A4J4W0 A0A2W1BG02 A0A1Y1N5S4 A0A1Y1N8Q1 A0A1Y1NAJ4 A0A1Y1N5N1 A0A1Y1N680 A0A1Y1NC98 A0A1Y1N719 A0A1Y1N9E0 A0A2J7R6J0 A0A2J7R6K3 A0A2J7R6J4 A0A2J7R6J7 D6WQX0 E0VAR1 A0A067QPR7 A0A0A9WVI4 A0A1Y1N5V1 N6SW71 A0A2S2PPT7 A0A2S2QXJ5 J9K9G0 A0A2J7R6J8 A0A2H8TN70 A0A2J7R6K8 E2B806 A0A154P8A3 A0A224XDS9 F4WP99 A0A195E5P1 A0A336LX67 A0A151WMJ0 A0A195EUJ2 A0A336LKL2 A0A336LPD6 A0A336LJW4 A0A336K0D9 A0A336K2W3 A0A026WNT8 A0A195C8C2 A0A0L7R928 A0A195B125 A0A0N8ES16 A0A1S4FZ82 Q16J05 A0A0C9QND1 A0A087ZX61 A0A310SCC2 B0WVU5 A0A0C9R5J4 K7IZC0 A0A1Q3EXQ7 A0A2M4BAJ0 A0A1Q3EXS2 A0A232EX52 A0A158NR65 A0A1Q3EXL2 A0A182FG24 A0A2M4A7J3 A0A2M4B994 A0A2M4B9A7 A0A2M4B961 W5JH27 E9J571 A0A182T4A1 A0A3L8DWX1 A0A182NHB7 A0A182QXC4 Q7QJX4 A0A1L8DLM0 E2ADU0 A0A084VF60 A0A182RB14 A0A182WIZ1 A0A182LWP7 V9IKV3 A0A0J7NI72 A0A182IM63 A0A0C9PM36 A0A182JQW3 A0A0A9VU05 A0A146MC82 W4VRK5
Pubmed
EMBL
RSAL01000029
RVE51749.1
KQ459166
KPJ03581.1
BABH01025308
BABH01025309
+ More
BABH01025310 NWSH01003367 PCG66454.1 PCG66450.1 PCG66452.1 PCG66451.1 PCG66449.1 PCG66453.1 KZ150264 PZC71710.1 GEZM01012049 JAV93243.1 GEZM01012052 JAV93240.1 GEZM01012046 JAV93246.1 GEZM01012050 JAV93242.1 GEZM01012053 JAV93239.1 GEZM01012045 JAV93247.1 GEZM01012044 JAV93248.1 GEZM01012047 JAV93245.1 NEVH01006756 PNF36455.1 PNF36450.1 PNF36449.1 PNF36454.1 KQ971351 EFA06518.2 DS235014 EEB10467.1 KK853153 KDR10554.1 GBHO01032168 JAG11436.1 GEZM01012051 JAV93241.1 APGK01054093 APGK01054094 APGK01054095 APGK01054096 KB741247 ENN72009.1 GGMR01018824 MBY31443.1 GGMS01013288 MBY82491.1 ABLF02036508 PNF36461.1 GFXV01002923 MBW14728.1 PNF36460.1 GL446268 EFN88191.1 KQ434844 KZC08155.1 GFTR01008478 JAW07948.1 GL888243 EGI64080.1 KQ979608 KYN20483.1 UFQS01000031 UFQT01000031 SSW97852.1 SSX18238.1 KQ982944 KYQ49021.1 KQ981965 KYN31826.1 SSW97849.1 SSX18235.1 SSW97851.1 SSX18237.1 SSW97850.1 SSX18236.1 SSW97847.1 SSX18233.1 SSW97848.1 SSX18234.1 KK107139 EZA57722.1 KQ978081 KYM97134.1 KQ414628 KOC67372.1 KQ976691 KYM77904.1 GDUN01000722 JAN95197.1 CH478040 EAT34249.1 GBYB01002152 JAG71919.1 KQ764513 OAD54508.1 DS232132 EDS35777.1 GBYB01008052 JAG77819.1 GFDL01014953 JAV20092.1 GGFJ01000906 MBW50047.1 GFDL01014936 JAV20109.1 NNAY01001776 OXU22951.1 ADTU01023750 ADTU01023751 ADTU01023752 ADTU01023753 GFDL01014996 JAV20049.1 GGFK01003404 MBW36725.1 GGFJ01000483 MBW49624.1 GGFJ01000465 MBW49606.1 GGFJ01000464 MBW49605.1 ADMH02001258 ETN63366.1 GL768124 EFZ12020.1 QOIP01000003 RLU24937.1 AXCN02000894 AAAB01008807 EAA04687.4 GFDF01006830 JAV07254.1 GL438820 EFN68452.1 ATLV01012362 KE524785 KFB36604.1 AXCM01000589 JR049845 AEY61096.1 LBMM01004653 KMQ92205.1 GBYB01002153 JAG71920.1 GBHO01044415 JAF99188.1 GDHC01002459 JAQ16170.1 GANO01002332 JAB57539.1
BABH01025310 NWSH01003367 PCG66454.1 PCG66450.1 PCG66452.1 PCG66451.1 PCG66449.1 PCG66453.1 KZ150264 PZC71710.1 GEZM01012049 JAV93243.1 GEZM01012052 JAV93240.1 GEZM01012046 JAV93246.1 GEZM01012050 JAV93242.1 GEZM01012053 JAV93239.1 GEZM01012045 JAV93247.1 GEZM01012044 JAV93248.1 GEZM01012047 JAV93245.1 NEVH01006756 PNF36455.1 PNF36450.1 PNF36449.1 PNF36454.1 KQ971351 EFA06518.2 DS235014 EEB10467.1 KK853153 KDR10554.1 GBHO01032168 JAG11436.1 GEZM01012051 JAV93241.1 APGK01054093 APGK01054094 APGK01054095 APGK01054096 KB741247 ENN72009.1 GGMR01018824 MBY31443.1 GGMS01013288 MBY82491.1 ABLF02036508 PNF36461.1 GFXV01002923 MBW14728.1 PNF36460.1 GL446268 EFN88191.1 KQ434844 KZC08155.1 GFTR01008478 JAW07948.1 GL888243 EGI64080.1 KQ979608 KYN20483.1 UFQS01000031 UFQT01000031 SSW97852.1 SSX18238.1 KQ982944 KYQ49021.1 KQ981965 KYN31826.1 SSW97849.1 SSX18235.1 SSW97851.1 SSX18237.1 SSW97850.1 SSX18236.1 SSW97847.1 SSX18233.1 SSW97848.1 SSX18234.1 KK107139 EZA57722.1 KQ978081 KYM97134.1 KQ414628 KOC67372.1 KQ976691 KYM77904.1 GDUN01000722 JAN95197.1 CH478040 EAT34249.1 GBYB01002152 JAG71919.1 KQ764513 OAD54508.1 DS232132 EDS35777.1 GBYB01008052 JAG77819.1 GFDL01014953 JAV20092.1 GGFJ01000906 MBW50047.1 GFDL01014936 JAV20109.1 NNAY01001776 OXU22951.1 ADTU01023750 ADTU01023751 ADTU01023752 ADTU01023753 GFDL01014996 JAV20049.1 GGFK01003404 MBW36725.1 GGFJ01000483 MBW49624.1 GGFJ01000465 MBW49606.1 GGFJ01000464 MBW49605.1 ADMH02001258 ETN63366.1 GL768124 EFZ12020.1 QOIP01000003 RLU24937.1 AXCN02000894 AAAB01008807 EAA04687.4 GFDF01006830 JAV07254.1 GL438820 EFN68452.1 ATLV01012362 KE524785 KFB36604.1 AXCM01000589 JR049845 AEY61096.1 LBMM01004653 KMQ92205.1 GBYB01002153 JAG71920.1 GBHO01044415 JAF99188.1 GDHC01002459 JAQ16170.1 GANO01002332 JAB57539.1
Proteomes
UP000283053
UP000053268
UP000005204
UP000218220
UP000235965
UP000007266
+ More
UP000009046 UP000027135 UP000019118 UP000007819 UP000008237 UP000076502 UP000007755 UP000078492 UP000075809 UP000078541 UP000053097 UP000078542 UP000053825 UP000078540 UP000008820 UP000005203 UP000002320 UP000002358 UP000215335 UP000005205 UP000069272 UP000000673 UP000075901 UP000279307 UP000075884 UP000075886 UP000007062 UP000000311 UP000030765 UP000075900 UP000075920 UP000075883 UP000036403 UP000075880 UP000075881
UP000009046 UP000027135 UP000019118 UP000007819 UP000008237 UP000076502 UP000007755 UP000078492 UP000075809 UP000078541 UP000053097 UP000078542 UP000053825 UP000078540 UP000008820 UP000005203 UP000002320 UP000002358 UP000215335 UP000005205 UP000069272 UP000000673 UP000075901 UP000279307 UP000075884 UP000075886 UP000007062 UP000000311 UP000030765 UP000075900 UP000075920 UP000075883 UP000036403 UP000075880 UP000075881
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A3S2LE36
A0A0N1IA73
H9JW58
A0A2A4J4L6
A0A2A4J4T9
A0A2A4J4X7
+ More
A0A2A4J373 A0A2A4J3K4 A0A2A4J4W0 A0A2W1BG02 A0A1Y1N5S4 A0A1Y1N8Q1 A0A1Y1NAJ4 A0A1Y1N5N1 A0A1Y1N680 A0A1Y1NC98 A0A1Y1N719 A0A1Y1N9E0 A0A2J7R6J0 A0A2J7R6K3 A0A2J7R6J4 A0A2J7R6J7 D6WQX0 E0VAR1 A0A067QPR7 A0A0A9WVI4 A0A1Y1N5V1 N6SW71 A0A2S2PPT7 A0A2S2QXJ5 J9K9G0 A0A2J7R6J8 A0A2H8TN70 A0A2J7R6K8 E2B806 A0A154P8A3 A0A224XDS9 F4WP99 A0A195E5P1 A0A336LX67 A0A151WMJ0 A0A195EUJ2 A0A336LKL2 A0A336LPD6 A0A336LJW4 A0A336K0D9 A0A336K2W3 A0A026WNT8 A0A195C8C2 A0A0L7R928 A0A195B125 A0A0N8ES16 A0A1S4FZ82 Q16J05 A0A0C9QND1 A0A087ZX61 A0A310SCC2 B0WVU5 A0A0C9R5J4 K7IZC0 A0A1Q3EXQ7 A0A2M4BAJ0 A0A1Q3EXS2 A0A232EX52 A0A158NR65 A0A1Q3EXL2 A0A182FG24 A0A2M4A7J3 A0A2M4B994 A0A2M4B9A7 A0A2M4B961 W5JH27 E9J571 A0A182T4A1 A0A3L8DWX1 A0A182NHB7 A0A182QXC4 Q7QJX4 A0A1L8DLM0 E2ADU0 A0A084VF60 A0A182RB14 A0A182WIZ1 A0A182LWP7 V9IKV3 A0A0J7NI72 A0A182IM63 A0A0C9PM36 A0A182JQW3 A0A0A9VU05 A0A146MC82 W4VRK5
A0A2A4J373 A0A2A4J3K4 A0A2A4J4W0 A0A2W1BG02 A0A1Y1N5S4 A0A1Y1N8Q1 A0A1Y1NAJ4 A0A1Y1N5N1 A0A1Y1N680 A0A1Y1NC98 A0A1Y1N719 A0A1Y1N9E0 A0A2J7R6J0 A0A2J7R6K3 A0A2J7R6J4 A0A2J7R6J7 D6WQX0 E0VAR1 A0A067QPR7 A0A0A9WVI4 A0A1Y1N5V1 N6SW71 A0A2S2PPT7 A0A2S2QXJ5 J9K9G0 A0A2J7R6J8 A0A2H8TN70 A0A2J7R6K8 E2B806 A0A154P8A3 A0A224XDS9 F4WP99 A0A195E5P1 A0A336LX67 A0A151WMJ0 A0A195EUJ2 A0A336LKL2 A0A336LPD6 A0A336LJW4 A0A336K0D9 A0A336K2W3 A0A026WNT8 A0A195C8C2 A0A0L7R928 A0A195B125 A0A0N8ES16 A0A1S4FZ82 Q16J05 A0A0C9QND1 A0A087ZX61 A0A310SCC2 B0WVU5 A0A0C9R5J4 K7IZC0 A0A1Q3EXQ7 A0A2M4BAJ0 A0A1Q3EXS2 A0A232EX52 A0A158NR65 A0A1Q3EXL2 A0A182FG24 A0A2M4A7J3 A0A2M4B994 A0A2M4B9A7 A0A2M4B961 W5JH27 E9J571 A0A182T4A1 A0A3L8DWX1 A0A182NHB7 A0A182QXC4 Q7QJX4 A0A1L8DLM0 E2ADU0 A0A084VF60 A0A182RB14 A0A182WIZ1 A0A182LWP7 V9IKV3 A0A0J7NI72 A0A182IM63 A0A0C9PM36 A0A182JQW3 A0A0A9VU05 A0A146MC82 W4VRK5
PDB
4D0N
E-value=4.66017e-48,
Score=485
Ontologies
GO
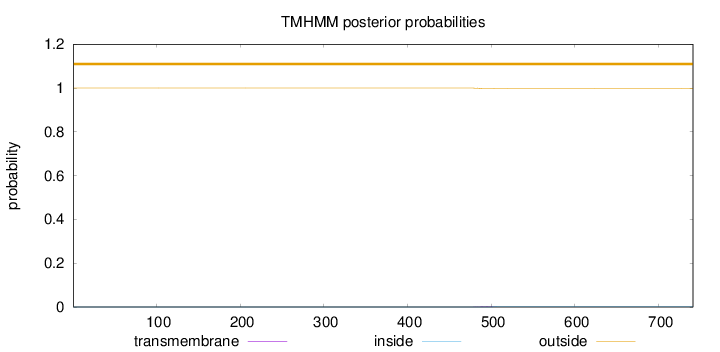
Topology
Length:
741
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03009
Exp number, first 60 AAs:
0.00014
Total prob of N-in:
0.00019
outside
1 - 741
Population Genetic Test Statistics
Pi
268.8299
Theta
179.063175
Tajima's D
1.492186
CLR
0.177008
CSRT
0.789310534473276
Interpretation
Uncertain
