Gene
KWMTBOMO16301
Annotation
PREDICTED:_uncharacterized_protein_LOC101739175_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.044 Nuclear Reliability : 1.494 PlasmaMembrane Reliability : 1.101
Sequence
CDS
ATGTCTCAAACATGGGACATTTTGCCACTACTTCCACTGAGGTGTGTGCTGGATTACCTGAGCATGGAGGATGCCTTGGCAGCAATGTCCACATGTAGGCATTGGAGGGATGCTGTTTTATTATATGAAGGATATAAGGAAACTGTAAAGCTAAACACAGGGAATTTAGAAAAGAATTTGTTTTTAACGCGCATGTTCAGAAGGAACGCTAGGAAACTACATATAGTCATCGAGAATGAAGTGGATTTAGACAAATTCGTCAGCTTCGTGTTACCACAATATTTCGATTCTGTGAAATTAACGGAACTAACGTTCGTCGGGCCCAGTTACGTGCGTGTCAACAAAATGCCCATTGTAAAATTGAAAAGGATAATAACCGAGAGTCTTATATTGAAACACGCGCATAGTTTACAGAGCTTATCGATATTAGGTTGTGAAATGGGCTTCGCCAAAAATGATAACGAGAGGTATCATAATAAACAAGTGGAGTATTTTTCGAGGTCTCTATCATTCAACACCGTCCCGTGTCCCGAGGACTCGGTCCTGTCCAGTAGGAACGTCAATCTGATGATGTTCTCATCGTTGCAGCATATCATATTAGATTACGAGCAGGTGAACAATCCGTCGTTGGAGACGCTCTCGAACCTGTCCAACTTCAATATGCTATCGCTGAACGTGATGTACAAGAGGAACATGAGGCTGAAGCGTCTGGACTGGCACCGAATCAACAGGGTCTTCGATAATAGATTGAATGTTGCCGTCAATATTATCACTACACCGTATAGGATGTTCTACGAATTGATGAACAACGTTCTAGTCGAGGGGATGTCTCTGTGTTCATTAAAGGTGTTTTTCTGCGAGGCCCTTCACACGCCGTTCCTGCATCATGTCGTCGATAAATTCCACCTGACGCTGCGGGAGCTCGTGTGGGTGGACTGCCCGAATGACTCAAACGATAAGTACGAGCGCCTCATCAGTTCCACGCACAACATTATTGCGGAGGACTCGATAAATGTGAATCCTTTCGTATTGCTCTGCTGGCAATGTGAACGATTGCAGAGACTCGTTATTCATGGGTACTGGATCTGGCAGTATGACGTCATCGGGTTCGTGCGGCTTCGCAAGTCGCTCCGGCAGCTGGACGTGTCGGCGGTGCACCGGCGCCGGGGCCGCGCCGCCGCCGGCCCGCCGCGCGTGCTGCTCGCCGACGTGGCCGACATCACCTCGCCCAGAGCCGTGCAGGTCGCTAGCCTCACAATACACGCCTCCCGTATCCCGGAGCTGAAGTGGCGGCCGACGCCGTGGGGGGCGCTGCACCCGGCGCTGCGCTCCCGCTCGCGCGCCAAGCAGCGCCTCGACTACATCCGCCGCGAGCTGGCGCGCCCCACCGCCTGCAGGTACAGAAAGGATCGAGTTGACCAAAGTTAA
Protein
MSQTWDILPLLPLRCVLDYLSMEDALAAMSTCRHWRDAVLLYEGYKETVKLNTGNLEKNLFLTRMFRRNARKLHIVIENEVDLDKFVSFVLPQYFDSVKLTELTFVGPSYVRVNKMPIVKLKRIITESLILKHAHSLQSLSILGCEMGFAKNDNERYHNKQVEYFSRSLSFNTVPCPEDSVLSSRNVNLMMFSSLQHIILDYEQVNNPSLETLSNLSNFNMLSLNVMYKRNMRLKRLDWHRINRVFDNRLNVAVNIITTPYRMFYELMNNVLVEGMSLCSLKVFFCEALHTPFLHHVVDKFHLTLRELVWVDCPNDSNDKYERLISSTHNIIAEDSINVNPFVLLCWQCERLQRLVIHGYWIWQYDVIGFVRLRKSLRQLDVSAVHRRRGRAAAGPPRVLLADVADITSPRAVQVASLTIHASRIPELKWRPTPWGALHPALRSRSRAKQRLDYIRRELARPTACRYRKDRVDQS
Summary
Uniprot
A0A3S2M5C0
A0A1E1W6J8
A0A2W1BTA3
A0A0N1I3F7
A0A0N0PD53
A0A0A1XAM8
+ More
A0A1B6C1Q6 A0A0K8UX13 N6SVN4 A0A034VGN4 W8AR46 A0A1W4XJH6 A0A0K8TXF6 A0A182Y180 A0A1A9XYP0 A0A1B0BRD0 A0A1A9ZF31 A0A0K8TL71 M9PEL5 Q9VST7 A0A3L8E5M1 A0A182MM16 A0A182PHK1 A0A0L0C4P8 Q7Q6X2 D6WPM8 A0A182LJ78 A0A1B6LRU6 B4QMP8 A0A1I8NZF6 A0A182WB27 B4HKK6 A0A2J7R3V6 A0A1W4VCQ5 A0A182RRV2 A0A182Q0D0 R7T5Q3 A0A1I8N7K3 A0A1B0GJL6 A0A232FDN6 Q29F57 B4HB09 A0A1B6FMI1 A0A1A9VEA5 A0A3B0KGB5 B3NFP9 A0A1Q3EXK6 B4PFE9 A0A0N0U6A8 A0A1A9W7L6 A0A026WLL6 B3MA42 A0A2M4ACM2 A0A2M4ACL4 B0WLR5 B4N6X6 A0A0M4EJ52 A0A151X996 E2BIL9 A0A087UBU3 K7ISF9 A0A154NZ65
A0A1B6C1Q6 A0A0K8UX13 N6SVN4 A0A034VGN4 W8AR46 A0A1W4XJH6 A0A0K8TXF6 A0A182Y180 A0A1A9XYP0 A0A1B0BRD0 A0A1A9ZF31 A0A0K8TL71 M9PEL5 Q9VST7 A0A3L8E5M1 A0A182MM16 A0A182PHK1 A0A0L0C4P8 Q7Q6X2 D6WPM8 A0A182LJ78 A0A1B6LRU6 B4QMP8 A0A1I8NZF6 A0A182WB27 B4HKK6 A0A2J7R3V6 A0A1W4VCQ5 A0A182RRV2 A0A182Q0D0 R7T5Q3 A0A1I8N7K3 A0A1B0GJL6 A0A232FDN6 Q29F57 B4HB09 A0A1B6FMI1 A0A1A9VEA5 A0A3B0KGB5 B3NFP9 A0A1Q3EXK6 B4PFE9 A0A0N0U6A8 A0A1A9W7L6 A0A026WLL6 B3MA42 A0A2M4ACM2 A0A2M4ACL4 B0WLR5 B4N6X6 A0A0M4EJ52 A0A151X996 E2BIL9 A0A087UBU3 K7ISF9 A0A154NZ65
Pubmed
28756777
26354079
25830018
23537049
25348373
24495485
+ More
25244985 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30249741 26108605 12364791 18362917 19820115 17994087 22936249 23254933 25315136 28648823 15632085 17550304 24508170 20798317 20075255
25244985 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30249741 26108605 12364791 18362917 19820115 17994087 22936249 23254933 25315136 28648823 15632085 17550304 24508170 20798317 20075255
EMBL
RSAL01000029
RVE51747.1
GDQN01008446
GDQN01008111
JAT82608.1
JAT82943.1
+ More
KZ149943 PZC76874.1 KQ459166 KPJ03583.1 KQ460460 KPJ14636.1 GBXI01013294 GBXI01006559 JAD00998.1 JAD07733.1 GEDC01029845 JAS07453.1 GDHF01023665 GDHF01021112 JAI28649.1 JAI31202.1 APGK01054913 KB741253 KB632340 ENN71814.1 ERL92956.1 GAKP01016471 GAKP01016470 JAC42481.1 GAMC01019543 GAMC01019542 JAB87013.1 GDHF01033584 JAI18730.1 JXJN01019046 GDAI01002675 JAI14928.1 AE014296 AGB94308.1 BT016122 AAF50324.1 AAV37007.1 AHN58015.1 QOIP01000001 RLU27399.1 AXCM01007282 JRES01000919 KNC27260.1 AAAB01008960 EAA11455.3 KQ971354 EFA06838.1 GEBQ01013554 JAT26423.1 CM000363 CM002912 EDX09802.1 KMY98552.1 CH480815 EDW40809.1 NEVH01007818 PNF35495.1 AXCN02000366 AMQN01015312 KB311780 ELT88615.1 AJWK01021977 AJWK01021978 AJWK01021979 NNAY01000364 OXU28871.1 CH379068 EAL31215.2 CH479246 EDW37803.1 GECZ01018353 JAS51416.1 OUUW01000009 SPP85379.1 CH954178 EDV50591.1 GFDL01015039 JAV20006.1 CM000159 EDW93075.1 KQ435732 KOX77394.1 KK107154 EZA56935.1 CH902618 EDV41262.1 GGFK01005198 MBW38519.1 GGFK01005213 MBW38534.1 DS231990 EDS30643.1 CH964168 EDW80115.1 CP012525 ALC44001.1 KQ982383 KYQ56904.1 GL448516 EFN84466.1 KK119125 KFM74832.1 KQ434783 KZC04969.1
KZ149943 PZC76874.1 KQ459166 KPJ03583.1 KQ460460 KPJ14636.1 GBXI01013294 GBXI01006559 JAD00998.1 JAD07733.1 GEDC01029845 JAS07453.1 GDHF01023665 GDHF01021112 JAI28649.1 JAI31202.1 APGK01054913 KB741253 KB632340 ENN71814.1 ERL92956.1 GAKP01016471 GAKP01016470 JAC42481.1 GAMC01019543 GAMC01019542 JAB87013.1 GDHF01033584 JAI18730.1 JXJN01019046 GDAI01002675 JAI14928.1 AE014296 AGB94308.1 BT016122 AAF50324.1 AAV37007.1 AHN58015.1 QOIP01000001 RLU27399.1 AXCM01007282 JRES01000919 KNC27260.1 AAAB01008960 EAA11455.3 KQ971354 EFA06838.1 GEBQ01013554 JAT26423.1 CM000363 CM002912 EDX09802.1 KMY98552.1 CH480815 EDW40809.1 NEVH01007818 PNF35495.1 AXCN02000366 AMQN01015312 KB311780 ELT88615.1 AJWK01021977 AJWK01021978 AJWK01021979 NNAY01000364 OXU28871.1 CH379068 EAL31215.2 CH479246 EDW37803.1 GECZ01018353 JAS51416.1 OUUW01000009 SPP85379.1 CH954178 EDV50591.1 GFDL01015039 JAV20006.1 CM000159 EDW93075.1 KQ435732 KOX77394.1 KK107154 EZA56935.1 CH902618 EDV41262.1 GGFK01005198 MBW38519.1 GGFK01005213 MBW38534.1 DS231990 EDS30643.1 CH964168 EDW80115.1 CP012525 ALC44001.1 KQ982383 KYQ56904.1 GL448516 EFN84466.1 KK119125 KFM74832.1 KQ434783 KZC04969.1
Proteomes
UP000283053
UP000053268
UP000053240
UP000019118
UP000030742
UP000192223
+ More
UP000076408 UP000092443 UP000092460 UP000092445 UP000000803 UP000279307 UP000075883 UP000075885 UP000037069 UP000007062 UP000007266 UP000000304 UP000095300 UP000075920 UP000001292 UP000235965 UP000192221 UP000075900 UP000075886 UP000014760 UP000095301 UP000092461 UP000215335 UP000001819 UP000008744 UP000078200 UP000268350 UP000008711 UP000002282 UP000053105 UP000091820 UP000053097 UP000007801 UP000002320 UP000007798 UP000092553 UP000075809 UP000008237 UP000054359 UP000002358 UP000076502
UP000076408 UP000092443 UP000092460 UP000092445 UP000000803 UP000279307 UP000075883 UP000075885 UP000037069 UP000007062 UP000007266 UP000000304 UP000095300 UP000075920 UP000001292 UP000235965 UP000192221 UP000075900 UP000075886 UP000014760 UP000095301 UP000092461 UP000215335 UP000001819 UP000008744 UP000078200 UP000268350 UP000008711 UP000002282 UP000053105 UP000091820 UP000053097 UP000007801 UP000002320 UP000007798 UP000092553 UP000075809 UP000008237 UP000054359 UP000002358 UP000076502
PRIDE
Interpro
SUPFAM
SSF81383
SSF81383
Gene 3D
ProteinModelPortal
A0A3S2M5C0
A0A1E1W6J8
A0A2W1BTA3
A0A0N1I3F7
A0A0N0PD53
A0A0A1XAM8
+ More
A0A1B6C1Q6 A0A0K8UX13 N6SVN4 A0A034VGN4 W8AR46 A0A1W4XJH6 A0A0K8TXF6 A0A182Y180 A0A1A9XYP0 A0A1B0BRD0 A0A1A9ZF31 A0A0K8TL71 M9PEL5 Q9VST7 A0A3L8E5M1 A0A182MM16 A0A182PHK1 A0A0L0C4P8 Q7Q6X2 D6WPM8 A0A182LJ78 A0A1B6LRU6 B4QMP8 A0A1I8NZF6 A0A182WB27 B4HKK6 A0A2J7R3V6 A0A1W4VCQ5 A0A182RRV2 A0A182Q0D0 R7T5Q3 A0A1I8N7K3 A0A1B0GJL6 A0A232FDN6 Q29F57 B4HB09 A0A1B6FMI1 A0A1A9VEA5 A0A3B0KGB5 B3NFP9 A0A1Q3EXK6 B4PFE9 A0A0N0U6A8 A0A1A9W7L6 A0A026WLL6 B3MA42 A0A2M4ACM2 A0A2M4ACL4 B0WLR5 B4N6X6 A0A0M4EJ52 A0A151X996 E2BIL9 A0A087UBU3 K7ISF9 A0A154NZ65
A0A1B6C1Q6 A0A0K8UX13 N6SVN4 A0A034VGN4 W8AR46 A0A1W4XJH6 A0A0K8TXF6 A0A182Y180 A0A1A9XYP0 A0A1B0BRD0 A0A1A9ZF31 A0A0K8TL71 M9PEL5 Q9VST7 A0A3L8E5M1 A0A182MM16 A0A182PHK1 A0A0L0C4P8 Q7Q6X2 D6WPM8 A0A182LJ78 A0A1B6LRU6 B4QMP8 A0A1I8NZF6 A0A182WB27 B4HKK6 A0A2J7R3V6 A0A1W4VCQ5 A0A182RRV2 A0A182Q0D0 R7T5Q3 A0A1I8N7K3 A0A1B0GJL6 A0A232FDN6 Q29F57 B4HB09 A0A1B6FMI1 A0A1A9VEA5 A0A3B0KGB5 B3NFP9 A0A1Q3EXK6 B4PFE9 A0A0N0U6A8 A0A1A9W7L6 A0A026WLL6 B3MA42 A0A2M4ACM2 A0A2M4ACL4 B0WLR5 B4N6X6 A0A0M4EJ52 A0A151X996 E2BIL9 A0A087UBU3 K7ISF9 A0A154NZ65
Ontologies
GO
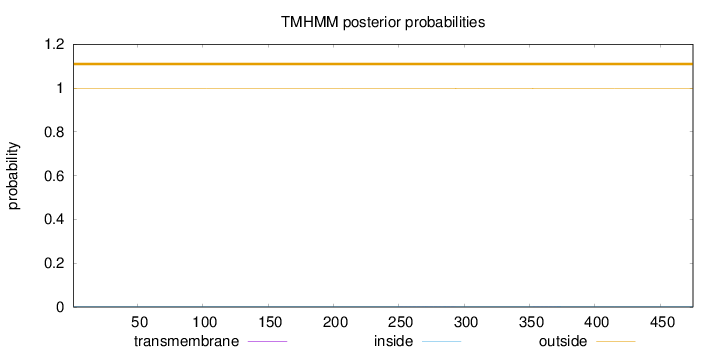
Topology
Length:
475
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02677
Exp number, first 60 AAs:
0.0022
Total prob of N-in:
0.00185
outside
1 - 475
Population Genetic Test Statistics
Pi
210.551053
Theta
150.811453
Tajima's D
1.293076
CLR
0.219156
CSRT
0.741462926853657
Interpretation
Uncertain
