Gene
KWMTBOMO16297
Pre Gene Modal
BGIBMGA002240
Annotation
PREDICTED:_uncharacterized_protein_LOC106717303_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 2.648
Sequence
CDS
ATGAAGCCTCTGTCAAAACTTGAAATGCAGTGTGAAGAGCATTTTAAGAAAACTCACACTCGAAACACTGACGGTCGATATGAGGTTCGCCTACCCTTCAAGGACGAAGACGCTCCAAATTTAGGCAACTCCAGACAACTTGCTTTGAAACGCTTACATCAGATGGAGAATAAGTTTAAGCGAAGACCCGAGTTTCACGAAGAATACAGCAAGTTCATGAGACAATATGAGTCACTCGGTCACATGGAGGTCGTCCCTGAAACAGAGAAGACGAATAGAGCTTATCACCTACCACATCATGCTGTTCTTCGTGCCTCGAGCCTCACTACAAAGTTGCGCGTAGTATTTGACGGAAGCGCTAAGCCTGTAGACGGAAACTCACTTAACGATGAGTTACTCATCGGCCCACCGCTGCAACAAGATATACGAGATTTAGTAACACGGTGGAGACAACATAAGTACTGCCTAGTAGCTGATATACAACACATGTACCGACAGATACTTATTTCAAAGCAAGACACTGATTACCAGAGAATCTTGTGGCGCGAGTCATCGGAAGACCCGATTAGAGAATATCGGTTACTTACCGTCACATATGGCAGTTCATGTGCTCCATATCTTGCTATAAGGACACTTCATCAACTTGCAGAGGACGAACGTGGAAAGTTTCCTGAAGCTGAACTTCTAAAAACTGATCTCTATATGGACGATTTGATGACAGGCTCATCTACAGAAGAAGACACTCAACGGCTTCAACGTCGCTTAACTGAACTGTTTAAACGTGGCGGTTTTCTTCTACACAAGTGGTCGTCTAATAGTGAAATAGTTTTAAATGAGATTCCCGATTCGATAAAAGCATTAAAAGGTTCAGTGAATATTAAAATGGATGATTCGGTGAAAGCTCTGGGTATAGCCTGGAAACCTCAAAGTGATATATTTGAACTGGTAGTTAACTTACCTCATGACAACGACGTTGTTACAAAACGAAGTGTGCTATCTGCGATAGCAAAAACTTTTGACCCTCTGGGTTGGCTAGCACCTTGCGTGATTGTGTTAAAAATGTTTATGCAGAAGTTGTGGCTGGCCGGCCTGGACTGGGACTCTGAACTTCCTGAAGACTTGAAAACTGAATGGCAGAATTACCTGACTAATTTTGAACACATGCAGGCCATTCAGCTTCCACGTTGGCTAGGGATTTCAAATAATGTCAAAATTGAATTGCACGGCTATTGTGACGCCTCTTGCGCCGCCTACGCTGCGGTTATTTACATCAGAGTAATCACGGACAATGAGATAAAAGTAAGTCTAGTCGCAGCCAAAACAAAGGTCGCTCCCGTTAAACAGATCTCATTGCCACGTCTTGAACTTTGTGGGGCAGTCTTATTGTCTAAATTGATACCATATGTCAAATCTAGTCTAAATATTGATGATAATTGTGTATTTGCTTGGACAGATTCCACAATAGTTTTGGCTTGGTTGCGTAAAACTCCAAATACTTGGAAGCCATTCGTTGGTAATCGCACTACCGAAATCTTGAATGTTACGAATAGCAGTCAATGGCATCATATTAAGTCTGCGGATAACCCCGCGGACTGCGCTTCACGCGGTATCAGTCCTGGCGAGCTGATGCAGTCACAACTATGGTGGGAAGGTCCAGCCTTTCTTCGCGAGTCTGTTGAGATTCGTTACCCTGATTTCACTATACCTGAAACCACACTCGAAGCAAAACCGAAAGCAAAAGTTAGTTACTTATGCACCTCTGAACCCAATGAATGTACTATGTACCTCAATAGATACTCAAATCTACTTAGACTTATACGTGTGACAGCCTATTGCTTTCGATTTTTGCGACTATGTAAAGATAAGGCTCTTAAAAGGAACGATAACATAAAACTGACATACCTCACTACTGATGAAATAAAAAATGCATTAAAAACTTGCATACGAATGTCACAGTCTGTTTACTTTAAAAGTGAAATAGGATTGTTATCACAGAGCAAACCTATCCCAAAAAATAGTAAACTTTTACCTTTGAATCCGATCCTAGACAATGAGAAAATTATTAGAGTCGGTGGACGTCTTGTAAATGCTCAAATACCACCTGATATGAAGTCTCCTATAATCTTACATCACAAATGTAACTTAGCAAAGTTGATTGTAATTGATGCTCATTTGAGAACACTACATGGTAGAAATTCGCTCACATTAAATGTTGTCCACCAAAAATATTGGATTCTCCGAGCAAAGAACTTAGTAAAAAAGATTTTAAGATTATGTAAACCTTGTTTTACCCAAACTGCACGACCTATGACTCCGTTAATGGGTAATTTACCACAGTGCAGAGTCAATCCGTCACGCCCGTTTTTGACCAGCGGCGTAGACTTTTGTGGTCCATTTACGCTGAAACTGTACTCTGGTAGATGCACCAAAACTTGTAAAGCTTACATAGGTGTGTTTTTATGTATGGTTACAAAGGCTATTCACCTTGAGCTGGTTAGTTCACTATCCAGTGCTGCGTTTTTAGCAGCCTTCAAACGTTTTACTTCCCGACGTGGGCATTGTAAAGACATCTGGAGTGATTGTGGGACTAACTTCATAGGCGCGTCCAAAGAATTGGATCTCGCCTTCAAAAACAGTAAATCCACTGTCGTTGACGAAATCGCCCAACTGTTAGCTAACGACAGCACAACATGGCATTTCATTCCCCCTGGTGCCCCTCACTTTGGAGGATTGTGGGAGGCAGGAGTTAAGTCCGTAAAAGGACATTTGAAACGTGTCGTAGGCGAGTCCTGCTTAACATTCGAAGAGTTTTGCACTCTGACCACACAGGTCGAGGCTTGTGTCAACTCTCGGCCTCTTACGTTGATTAGTTCTGCCGTTGATGAACTCCCGTTGACTCCTGGTCATTTTTTGATCGGCGAAGCACCGGTTACTATCCCTGAAGAAAGTTTCGTGGATACTCGACCTGACTATTTAAACCGATGGCAAAAGGTGCAACGTATGGTACAAAGTCTGTGGCGACGCTGGCAATCTGAGTATCTTACTATGTTGCAACATCGATACAAATGGTCCCAGAACCACCCCGACATAAATGTTGGTACGGTTGTCTTAGTGAAAGATGATCGTTTACCTCCAGGTAAATGGCTTTTGGGACGAATCGTGGCGAAGCACCCGGGAGCAGACGGTGTAACGCGTGTAGTCTCCTTACAATTTAAGGATCGTGTATTTAAGAGACCAGTTACTAAACTGTGCCCTCTGCCTCTAGCTGCTTCAGAACCTTAG
Protein
MKPLSKLEMQCEEHFKKTHTRNTDGRYEVRLPFKDEDAPNLGNSRQLALKRLHQMENKFKRRPEFHEEYSKFMRQYESLGHMEVVPETEKTNRAYHLPHHAVLRASSLTTKLRVVFDGSAKPVDGNSLNDELLIGPPLQQDIRDLVTRWRQHKYCLVADIQHMYRQILISKQDTDYQRILWRESSEDPIREYRLLTVTYGSSCAPYLAIRTLHQLAEDERGKFPEAELLKTDLYMDDLMTGSSTEEDTQRLQRRLTELFKRGGFLLHKWSSNSEIVLNEIPDSIKALKGSVNIKMDDSVKALGIAWKPQSDIFELVVNLPHDNDVVTKRSVLSAIAKTFDPLGWLAPCVIVLKMFMQKLWLAGLDWDSELPEDLKTEWQNYLTNFEHMQAIQLPRWLGISNNVKIELHGYCDASCAAYAAVIYIRVITDNEIKVSLVAAKTKVAPVKQISLPRLELCGAVLLSKLIPYVKSSLNIDDNCVFAWTDSTIVLAWLRKTPNTWKPFVGNRTTEILNVTNSSQWHHIKSADNPADCASRGISPGELMQSQLWWEGPAFLRESVEIRYPDFTIPETTLEAKPKAKVSYLCTSEPNECTMYLNRYSNLLRLIRVTAYCFRFLRLCKDKALKRNDNIKLTYLTTDEIKNALKTCIRMSQSVYFKSEIGLLSQSKPIPKNSKLLPLNPILDNEKIIRVGGRLVNAQIPPDMKSPIILHHKCNLAKLIVIDAHLRTLHGRNSLTLNVVHQKYWILRAKNLVKKILRLCKPCFTQTARPMTPLMGNLPQCRVNPSRPFLTSGVDFCGPFTLKLYSGRCTKTCKAYIGVFLCMVTKAIHLELVSSLSSAAFLAAFKRFTSRRGHCKDIWSDCGTNFIGASKELDLAFKNSKSTVVDEIAQLLANDSTTWHFIPPGAPHFGGLWEAGVKSVKGHLKRVVGESCLTFEEFCTLTTQVEACVNSRPLTLISSAVDELPLTPGHFLIGEAPVTIPEESFVDTRPDYLNRWQKVQRMVQSLWRRWQSEYLTMLQHRYKWSQNHPDINVGTVVLVKDDRLPPGKWLLGRIVAKHPGADGVTRVVSLQFKDRVFKRPVTKLCPLPLAASEP
Summary
Uniprot
A0A2H1WH28
A0A0N0PAG9
A0A0N1IHU1
A0A0J7KGI8
A0A226EPI5
A0A0J7KCR4
+ More
A0A1Y1LBZ3 A0A226E226 A0A226EUF5 A0A226DMG9 A0A0J7N5C9 A0A0J7N536 A0A0J7KB34 A0A226CWA5 A0A0J7K7L1 A0A0J7KCI2 A0A0J7N890 A0A0J7KFC5 A0A087T9T6 A0A226D417 A0A0J7ND98 A0A0J7KBZ0 A0A0J7N465 A0A2A4K6T9 A0A0J7KCB9 A0A0J7KBS8 A0A226E0X4 A0A0J7KJY9 A0A226EM86 A0A226EUK6 A0A226D0T3 W8CCT3 Q8I7Q1 A0A0J7KNK6 A0A069DYA1 J9LHB7 A0A2S2NCU7 A0A224XG39 X1WN86 A0A1Y1MTT1 A0A023F0J1 A0A0J7KBV8 A0A182H647 A0A182HA33 A0A1Y1N4E1 J9JNP4 X1WNM7 A0A0J7K9X4 A0A226EBC2 A0A023EZK8 A0A2S2PTX9 J9KCB6 A0A226DKA3 A0A1S3DIE2 A0A3Q0JDB5 A0A034W2D1 A0A226DUQ5 E9J9Y8 A0A0J7K9A7 A0A2S2QPS5 A0A0J7KCX3 J9KLD6 A0A3Q0JI79 J9JUK4 A0A2S2P1P0 A0A1S3DHZ9 A0A2S2Q6Z5 A0A1Y1L369 A7XDH0 A0A3L8DXG3 J9KD60 A0A2S2PNZ4 V9GZW9 X1WK09 A0A2S2NLW5 A0A1U8N896 Q4EBB8 A0A2M4CSC7 A0A2S2P963 A0A1W7R6F4 A0A2S2QLL5 Q8MNR4 X1WV59 J9JT64 J9K351 A0A0A9YD88 J9L9H5 A0A1X7UAA5 J9JUM9 W4VR47 Q4JS97 A0A2S2PNC2 A0A1Y1MY22 A0A1W7R6J2 A0A1W7R6L7 V5GHM7 A0A2S2PBK9 A0A1B6MUZ9 A0A0A9VSF7 A0A1Y1MYB7
A0A1Y1LBZ3 A0A226E226 A0A226EUF5 A0A226DMG9 A0A0J7N5C9 A0A0J7N536 A0A0J7KB34 A0A226CWA5 A0A0J7K7L1 A0A0J7KCI2 A0A0J7N890 A0A0J7KFC5 A0A087T9T6 A0A226D417 A0A0J7ND98 A0A0J7KBZ0 A0A0J7N465 A0A2A4K6T9 A0A0J7KCB9 A0A0J7KBS8 A0A226E0X4 A0A0J7KJY9 A0A226EM86 A0A226EUK6 A0A226D0T3 W8CCT3 Q8I7Q1 A0A0J7KNK6 A0A069DYA1 J9LHB7 A0A2S2NCU7 A0A224XG39 X1WN86 A0A1Y1MTT1 A0A023F0J1 A0A0J7KBV8 A0A182H647 A0A182HA33 A0A1Y1N4E1 J9JNP4 X1WNM7 A0A0J7K9X4 A0A226EBC2 A0A023EZK8 A0A2S2PTX9 J9KCB6 A0A226DKA3 A0A1S3DIE2 A0A3Q0JDB5 A0A034W2D1 A0A226DUQ5 E9J9Y8 A0A0J7K9A7 A0A2S2QPS5 A0A0J7KCX3 J9KLD6 A0A3Q0JI79 J9JUK4 A0A2S2P1P0 A0A1S3DHZ9 A0A2S2Q6Z5 A0A1Y1L369 A7XDH0 A0A3L8DXG3 J9KD60 A0A2S2PNZ4 V9GZW9 X1WK09 A0A2S2NLW5 A0A1U8N896 Q4EBB8 A0A2M4CSC7 A0A2S2P963 A0A1W7R6F4 A0A2S2QLL5 Q8MNR4 X1WV59 J9JT64 J9K351 A0A0A9YD88 J9L9H5 A0A1X7UAA5 J9JUM9 W4VR47 Q4JS97 A0A2S2PNC2 A0A1Y1MY22 A0A1W7R6J2 A0A1W7R6L7 V5GHM7 A0A2S2PBK9 A0A1B6MUZ9 A0A0A9VSF7 A0A1Y1MYB7
Pubmed
EMBL
ODYU01008608
SOQ52337.1
KQ458473
KPJ05885.1
KQ460036
KPJ18393.1
+ More
LBMM01007859 KMQ89359.1 LNIX01000002 OXA59522.1 LBMM01009551 KMQ88039.1 GEZM01060254 JAV71154.1 LNIX01000007 OXA51795.1 OXA60848.1 LNIX01000016 OXA46198.1 LBMM01009746 KMQ87900.1 LBMM01009898 KMQ87805.1 LBMM01010405 KMQ87487.1 LNIX01000069 OXA36894.1 LBMM01012188 KMQ86362.1 LBMM01009688 KMQ87939.1 LBMM01008464 KMQ88855.1 LBMM01008324 KMQ88972.1 KK114194 KFM61875.1 LNIX01000036 OXA39969.1 LBMM01006539 KMQ90520.1 LBMM01009964 KMQ87756.1 LBMM01010417 KMQ87475.1 NWSH01000070 PCG79985.1 LBMM01009645 KMQ87967.1 LBMM01009888 KMQ87813.1 OXA51382.1 LBMM01006490 KMQ90572.1 LNIX01000003 OXA58579.1 LNIX01000001 OXA61293.1 LNIX01000044 OXA38660.1 GAMC01003146 JAC03410.1 AY180917 AAN87269.1 LBMM01005079 KMQ91831.1 GBGD01000063 JAC88826.1 ABLF02022040 ABLF02056888 GGMR01002340 MBY14959.1 GFTR01008874 JAW07552.1 ABLF02041590 GEZM01021505 JAV88969.1 GBBI01004211 JAC14501.1 LBMM01009941 KMQ87772.1 JXUM01113346 KQ565681 KXJ70776.1 JXUM01030959 KQ560903 KXJ80414.1 GEZM01013098 JAV92753.1 ABLF02005684 ABLF02042806 LBMM01010986 KMQ87097.1 LNIX01000005 OXA54454.1 GBBI01004032 JAC14680.1 GGMR01020322 MBY32941.1 ABLF02042264 LNIX01000018 OXA45284.1 GAKP01010103 JAC48849.1 LNIX01000011 OXA48427.1 GL769615 EFZ10365.1 LBMM01011283 KMQ86917.1 GGMS01010524 MBY79727.1 LBMM01009210 KMQ88303.1 ABLF02013940 ABLF02036313 ABLF02066938 ABLF02066950 ABLF02039606 GGMR01010750 MBY23369.1 GGMS01004301 MBY73504.1 GEZM01066272 JAV68024.1 EF426679 ABP48078.1 QOIP01000003 RLU24775.1 ABLF02041863 GGMR01018506 MBY31125.1 U23420 AAB03640.1 ABLF02065856 GGMR01005535 MBY18154.1 AAGB01000056 EAL58741.1 GGFL01004049 MBW68227.1 GGMR01013370 MBY25989.1 GEHC01000898 JAV46747.1 GGMS01009443 MBY78646.1 AJ487856 CAD32253.1 ABLF02041731 ABLF02023842 ABLF02028949 ABLF02058391 ABLF02025193 GBHO01020765 GBHO01014531 JAG22839.1 JAG29073.1 ABLF02008636 ABLF02065931 ABLF02004035 ABLF02004036 ABLF02004037 ABLF02004038 ABLF02008018 ABLF02008020 ABLF02012635 ABLF02061171 GANO01004788 JAB55083.1 CR954257 CAJ14165.1 GGMR01018294 MBY30913.1 GEZM01018702 JAV90048.1 GEHC01000894 JAV46751.1 GEHC01000864 JAV46781.1 GALX01004947 JAB63519.1 GGMR01014165 MBY26784.1 GEBQ01007199 GEBQ01000222 JAT32778.1 JAT39755.1 GBHO01044317 GBHO01044315 JAF99286.1 JAF99288.1 GEZM01018619 JAV90138.1
LBMM01007859 KMQ89359.1 LNIX01000002 OXA59522.1 LBMM01009551 KMQ88039.1 GEZM01060254 JAV71154.1 LNIX01000007 OXA51795.1 OXA60848.1 LNIX01000016 OXA46198.1 LBMM01009746 KMQ87900.1 LBMM01009898 KMQ87805.1 LBMM01010405 KMQ87487.1 LNIX01000069 OXA36894.1 LBMM01012188 KMQ86362.1 LBMM01009688 KMQ87939.1 LBMM01008464 KMQ88855.1 LBMM01008324 KMQ88972.1 KK114194 KFM61875.1 LNIX01000036 OXA39969.1 LBMM01006539 KMQ90520.1 LBMM01009964 KMQ87756.1 LBMM01010417 KMQ87475.1 NWSH01000070 PCG79985.1 LBMM01009645 KMQ87967.1 LBMM01009888 KMQ87813.1 OXA51382.1 LBMM01006490 KMQ90572.1 LNIX01000003 OXA58579.1 LNIX01000001 OXA61293.1 LNIX01000044 OXA38660.1 GAMC01003146 JAC03410.1 AY180917 AAN87269.1 LBMM01005079 KMQ91831.1 GBGD01000063 JAC88826.1 ABLF02022040 ABLF02056888 GGMR01002340 MBY14959.1 GFTR01008874 JAW07552.1 ABLF02041590 GEZM01021505 JAV88969.1 GBBI01004211 JAC14501.1 LBMM01009941 KMQ87772.1 JXUM01113346 KQ565681 KXJ70776.1 JXUM01030959 KQ560903 KXJ80414.1 GEZM01013098 JAV92753.1 ABLF02005684 ABLF02042806 LBMM01010986 KMQ87097.1 LNIX01000005 OXA54454.1 GBBI01004032 JAC14680.1 GGMR01020322 MBY32941.1 ABLF02042264 LNIX01000018 OXA45284.1 GAKP01010103 JAC48849.1 LNIX01000011 OXA48427.1 GL769615 EFZ10365.1 LBMM01011283 KMQ86917.1 GGMS01010524 MBY79727.1 LBMM01009210 KMQ88303.1 ABLF02013940 ABLF02036313 ABLF02066938 ABLF02066950 ABLF02039606 GGMR01010750 MBY23369.1 GGMS01004301 MBY73504.1 GEZM01066272 JAV68024.1 EF426679 ABP48078.1 QOIP01000003 RLU24775.1 ABLF02041863 GGMR01018506 MBY31125.1 U23420 AAB03640.1 ABLF02065856 GGMR01005535 MBY18154.1 AAGB01000056 EAL58741.1 GGFL01004049 MBW68227.1 GGMR01013370 MBY25989.1 GEHC01000898 JAV46747.1 GGMS01009443 MBY78646.1 AJ487856 CAD32253.1 ABLF02041731 ABLF02023842 ABLF02028949 ABLF02058391 ABLF02025193 GBHO01020765 GBHO01014531 JAG22839.1 JAG29073.1 ABLF02008636 ABLF02065931 ABLF02004035 ABLF02004036 ABLF02004037 ABLF02004038 ABLF02008018 ABLF02008020 ABLF02012635 ABLF02061171 GANO01004788 JAB55083.1 CR954257 CAJ14165.1 GGMR01018294 MBY30913.1 GEZM01018702 JAV90048.1 GEHC01000894 JAV46751.1 GEHC01000864 JAV46781.1 GALX01004947 JAB63519.1 GGMR01014165 MBY26784.1 GEBQ01007199 GEBQ01000222 JAT32778.1 JAT39755.1 GBHO01044317 GBHO01044315 JAF99286.1 JAF99288.1 GEZM01018619 JAV90138.1
Proteomes
Pfam
Interpro
IPR001584
Integrase_cat-core
+ More
IPR012337 RNaseH-like_sf
IPR008042 Retrotrans_Pao
IPR001878 Znf_CCHC
IPR008737 Peptidase_asp_put
IPR040676 DUF5641
IPR005312 DUF1759
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR000477 RT_dom
IPR001995 Peptidase_A2_cat
IPR000727 T_SNARE_dom
IPR041373 RT_RNaseH
IPR001969 Aspartic_peptidase_AS
IPR041577 RT_RNaseH_2
IPR000425 MIP
IPR023271 Aquaporin-like
IPR022048 Envelope_fusion-like
IPR000331 Rap_GAP_dom
IPR035974 Rap/Ran-GAP_sf
IPR013087 Znf_C2H2_type
IPR012337 RNaseH-like_sf
IPR008042 Retrotrans_Pao
IPR001878 Znf_CCHC
IPR008737 Peptidase_asp_put
IPR040676 DUF5641
IPR005312 DUF1759
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR000477 RT_dom
IPR001995 Peptidase_A2_cat
IPR000727 T_SNARE_dom
IPR041373 RT_RNaseH
IPR001969 Aspartic_peptidase_AS
IPR041577 RT_RNaseH_2
IPR000425 MIP
IPR023271 Aquaporin-like
IPR022048 Envelope_fusion-like
IPR000331 Rap_GAP_dom
IPR035974 Rap/Ran-GAP_sf
IPR013087 Znf_C2H2_type
ProteinModelPortal
A0A2H1WH28
A0A0N0PAG9
A0A0N1IHU1
A0A0J7KGI8
A0A226EPI5
A0A0J7KCR4
+ More
A0A1Y1LBZ3 A0A226E226 A0A226EUF5 A0A226DMG9 A0A0J7N5C9 A0A0J7N536 A0A0J7KB34 A0A226CWA5 A0A0J7K7L1 A0A0J7KCI2 A0A0J7N890 A0A0J7KFC5 A0A087T9T6 A0A226D417 A0A0J7ND98 A0A0J7KBZ0 A0A0J7N465 A0A2A4K6T9 A0A0J7KCB9 A0A0J7KBS8 A0A226E0X4 A0A0J7KJY9 A0A226EM86 A0A226EUK6 A0A226D0T3 W8CCT3 Q8I7Q1 A0A0J7KNK6 A0A069DYA1 J9LHB7 A0A2S2NCU7 A0A224XG39 X1WN86 A0A1Y1MTT1 A0A023F0J1 A0A0J7KBV8 A0A182H647 A0A182HA33 A0A1Y1N4E1 J9JNP4 X1WNM7 A0A0J7K9X4 A0A226EBC2 A0A023EZK8 A0A2S2PTX9 J9KCB6 A0A226DKA3 A0A1S3DIE2 A0A3Q0JDB5 A0A034W2D1 A0A226DUQ5 E9J9Y8 A0A0J7K9A7 A0A2S2QPS5 A0A0J7KCX3 J9KLD6 A0A3Q0JI79 J9JUK4 A0A2S2P1P0 A0A1S3DHZ9 A0A2S2Q6Z5 A0A1Y1L369 A7XDH0 A0A3L8DXG3 J9KD60 A0A2S2PNZ4 V9GZW9 X1WK09 A0A2S2NLW5 A0A1U8N896 Q4EBB8 A0A2M4CSC7 A0A2S2P963 A0A1W7R6F4 A0A2S2QLL5 Q8MNR4 X1WV59 J9JT64 J9K351 A0A0A9YD88 J9L9H5 A0A1X7UAA5 J9JUM9 W4VR47 Q4JS97 A0A2S2PNC2 A0A1Y1MY22 A0A1W7R6J2 A0A1W7R6L7 V5GHM7 A0A2S2PBK9 A0A1B6MUZ9 A0A0A9VSF7 A0A1Y1MYB7
A0A1Y1LBZ3 A0A226E226 A0A226EUF5 A0A226DMG9 A0A0J7N5C9 A0A0J7N536 A0A0J7KB34 A0A226CWA5 A0A0J7K7L1 A0A0J7KCI2 A0A0J7N890 A0A0J7KFC5 A0A087T9T6 A0A226D417 A0A0J7ND98 A0A0J7KBZ0 A0A0J7N465 A0A2A4K6T9 A0A0J7KCB9 A0A0J7KBS8 A0A226E0X4 A0A0J7KJY9 A0A226EM86 A0A226EUK6 A0A226D0T3 W8CCT3 Q8I7Q1 A0A0J7KNK6 A0A069DYA1 J9LHB7 A0A2S2NCU7 A0A224XG39 X1WN86 A0A1Y1MTT1 A0A023F0J1 A0A0J7KBV8 A0A182H647 A0A182HA33 A0A1Y1N4E1 J9JNP4 X1WNM7 A0A0J7K9X4 A0A226EBC2 A0A023EZK8 A0A2S2PTX9 J9KCB6 A0A226DKA3 A0A1S3DIE2 A0A3Q0JDB5 A0A034W2D1 A0A226DUQ5 E9J9Y8 A0A0J7K9A7 A0A2S2QPS5 A0A0J7KCX3 J9KLD6 A0A3Q0JI79 J9JUK4 A0A2S2P1P0 A0A1S3DHZ9 A0A2S2Q6Z5 A0A1Y1L369 A7XDH0 A0A3L8DXG3 J9KD60 A0A2S2PNZ4 V9GZW9 X1WK09 A0A2S2NLW5 A0A1U8N896 Q4EBB8 A0A2M4CSC7 A0A2S2P963 A0A1W7R6F4 A0A2S2QLL5 Q8MNR4 X1WV59 J9JT64 J9K351 A0A0A9YD88 J9L9H5 A0A1X7UAA5 J9JUM9 W4VR47 Q4JS97 A0A2S2PNC2 A0A1Y1MY22 A0A1W7R6J2 A0A1W7R6L7 V5GHM7 A0A2S2PBK9 A0A1B6MUZ9 A0A0A9VSF7 A0A1Y1MYB7
Ontologies
GO
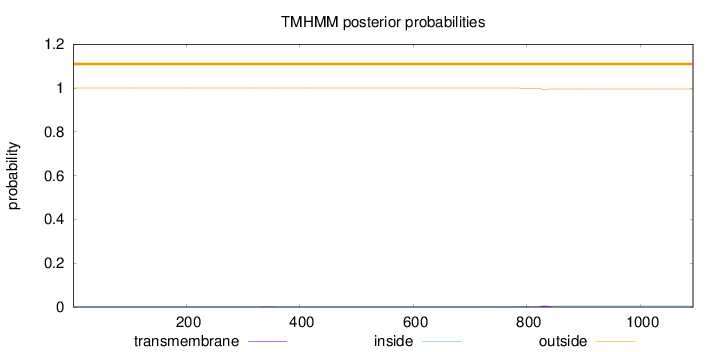
Topology
Length:
1093
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.12191
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00034
outside
1 - 1093
Population Genetic Test Statistics
Pi
376.218194
Theta
192.866422
Tajima's D
3.086891
CLR
0.381607
CSRT
0.983100844957752
Interpretation
Uncertain
