Pre Gene Modal
BGIBMGA014007
Annotation
hypothetical_protein_KGM_12813_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 3.242
Sequence
CDS
ATGGCATCTACATCCAATGATAACGTGCCTCAACATATGAATGCTCCAGAAAATGCTTCTGATGAACTTGAACCGGGTGCATGTCATGATAGCATACTAAACCCCACTGATTCCTTTGAAGAGTTTGCATCGGAGATGAGTTCCAGCCTAAGAGAGACCTGCTTGACTACTAAGAGCAGTACTATAAGTGAAATACAAAGTGAAATCTTGGAAACTGCTGAAGACCCTAAACTGAAAGCAGCGAAATCAAGTGATGATTTACAAACAAAAGATGGATCAATGTCAGCTTCAGCCAGCAATTCGAACTTAATTGAGGAGAATGGAGATGATACAGATGATGATAAAGACTATCTACAGTCACCAGAACTTGTTAATAAAGAGAAACATGTGTTCATACTTAGCACTGCAGGAAAACCTATTTATTCAAGATATGGCAACGAAGACAAGTTAGCTGGCCTGTGTGGTGTTATCCAAGCTCTTGTCAGCGTAATAGAAGAACAGAACCAGGATATATTGAGGTTCATCAACACGAAAGATTGTAAAGCAGTATTCCTTGTTAAGGGACCCCTAATACTGGTTGCTGTTTCGAAATCCAATGAAAGTGAAACCCAATTAGTTTTGCAGTTGACATATGCATTCAATCAAATTGTTTCGCTGTTAACACTAACCCAACTGAATAGAATATTTGAGCAGCGAAGGAACTATGATCTGAGAAGGCTACTGGCCGGAGCTGAAAGATTGATTGATCATTTGCTTGTGTTTATGGAGAAAGATCCTGCCTTTTTACTAGGTGCAGTCCGATGCCTTCCTTTACCAGAAAAAGCCAGAGAGAATATCACGAATGCAATAACAACCACGTGCCACAAGATTCGTGATCTCGTCTTCGCTATAATGATAGCTGGGAATCAACTGATAACATTAGTGAGAATGAAAAAATACACCCTGCACCCTTCGGATATACATTTGCTTTTCAATTTAATAAGGAGTTCGGAGTCATTCAAAACGGCCGAGAGCTGGACGCCAATATGCCTTCCCAAGTTTGATGCAACGGGCTTCCTGCACGGCCACGTGTCTTACATATCAGAGGACTGCCAGGCCTGTCTACTTCTACTGACCGTACAAAGGGATGCATTTACAACTTTATCCCAAGCCAAACACACAATAGCCGAGAAAATGAGACGGGCAAACTGTTTAGTTGCCATTAACAATGCTTTAAACCGTAACGAAGAGTTGTCCACAACGAAACCCCTCAAGCACATCGGCATACCGGAAATAAGGCATTTTATGTATAAATGTAAATCAACAGCGCAGCTGTTCTCGTCGGATCCGATTAGTTTAGATCGATTACAAGATTATCGATTAAGGCACAAAGAAGGGGGCGATATAGTCGATAAGAAAATAACAAAGTTGTCGGATATCGATTATGTTAGAAACTATCGGAAGTTTTGTAATCAGATACATTGTGTCGCGACGCCTGCTAAGCTTATATTCAGATCGAGTTCACAAGACACAATATTGTCTTGGGTGACAAACGGTTTCGAATTGTACGTGACGTTCGATCCGCTTATGGAGAAGGATCACGCGATCAAGGCCGTCGACCGACTCCTCAGGTGGATAAAGAAAGAAGAGGAGAGGATATTCATAATGAACGCTCCGACCTTCTAG
Protein
MASTSNDNVPQHMNAPENASDELEPGACHDSILNPTDSFEEFASEMSSSLRETCLTTKSSTISEIQSEILETAEDPKLKAAKSSDDLQTKDGSMSASASNSNLIEENGDDTDDDKDYLQSPELVNKEKHVFILSTAGKPIYSRYGNEDKLAGLCGVIQALVSVIEEQNQDILRFINTKDCKAVFLVKGPLILVAVSKSNESETQLVLQLTYAFNQIVSLLTLTQLNRIFEQRRNYDLRRLLAGAERLIDHLLVFMEKDPAFLLGAVRCLPLPEKARENITNAITTTCHKIRDLVFAIMIAGNQLITLVRMKKYTLHPSDIHLLFNLIRSSESFKTAESWTPICLPKFDATGFLHGHVSYISEDCQACLLLLTVQRDAFTTLSQAKHTIAEKMRRANCLVAINNALNRNEELSTTKPLKHIGIPEIRHFMYKCKSTAQLFSSDPISLDRLQDYRLRHKEGGDIVDKKITKLSDIDYVRNYRKFCNQIHCVATPAKLIFRSSSQDTILSWVTNGFELYVTFDPLMEKDHAIKAVDRLLRWIKKEEERIFIMNAPTF
Summary
Uniprot
H9JWU3
A0A2W1BKJ9
A0A2A4K7G1
A0A3S2NRA8
A0A212ET21
A0A2H1V351
+ More
A0A194Q2W4 A0A1E1WTH9 A0A0N0PDA4 A0A1E1W2B9 A0A2J7Q9G0 A0A067RTL3 D6WU67 A0A195FCH7 A0A158NBQ7 A0A195BF61 A0A195DR18 F4X862 A0A151X9C3 A0A087ZZR2 A0A2A3EQX7 A0A232EE22 A0A026WDC2 A0A151IKJ4 E2BC99 K7IUN0 E9IFJ1 E2A1P7 A0A154PI96 A0A0L7R1M9 A0A1W4WY10 A0A3L8DVP5 A0A2J7Q9G6 A0A1Y1LXA3 A0A1B6KRU6 A0A1S3H631 A0A182HRF5 A0A182KAU8 A0A182F1G2 A0A182RRA7 A0A182UTG5 W5JVP3 A0A182TUG4 A0A182PG39 A0A084W8R5 A0A182W8Y3 A0A182MTV1 A0A182QTR3 A0A182KYX7 Q7Q176 A0A182X2I9 A0A1B6K7Z4 R7TQ94 A0A182J4D9 A0A182NAU7 A0A1L8DB20 A0A1B0DG70 A0A1B0CHN4 Q16WP3 A0A1B0GQ66 T1IV58 A0A1Q3F345 B0W9V5 A0A1Q3F2X6 A0A182H047 N6SZB5 A0A0L8HD68 C3YLH1 A0A0K8TQM6 A0A2T7NTN1 A0A336MLY6 A0A087UF34 A0A0P4WJ08 A0A2B4SR23 A0A2C9K2S5 A0A0J7L2U0 A0A182YMP9 V4CBN3 A0A0P4VMH2 A0A0V0G670 T2MAS3 A0A1D2MUG4 A0A210PFP2 A0A0B6ZQH4 K1QH18 A0A224XR85 A7SAR3 Q9VR38 A0A0J9TFN7 B4NZ20 A0A0A1WVC6 A0A2H8TRD8 Q8MRZ0 V5HJG2 B4I174 B4N0J8 A0A034WHA9 B3N4A8 A0A0K2T5W3
A0A194Q2W4 A0A1E1WTH9 A0A0N0PDA4 A0A1E1W2B9 A0A2J7Q9G0 A0A067RTL3 D6WU67 A0A195FCH7 A0A158NBQ7 A0A195BF61 A0A195DR18 F4X862 A0A151X9C3 A0A087ZZR2 A0A2A3EQX7 A0A232EE22 A0A026WDC2 A0A151IKJ4 E2BC99 K7IUN0 E9IFJ1 E2A1P7 A0A154PI96 A0A0L7R1M9 A0A1W4WY10 A0A3L8DVP5 A0A2J7Q9G6 A0A1Y1LXA3 A0A1B6KRU6 A0A1S3H631 A0A182HRF5 A0A182KAU8 A0A182F1G2 A0A182RRA7 A0A182UTG5 W5JVP3 A0A182TUG4 A0A182PG39 A0A084W8R5 A0A182W8Y3 A0A182MTV1 A0A182QTR3 A0A182KYX7 Q7Q176 A0A182X2I9 A0A1B6K7Z4 R7TQ94 A0A182J4D9 A0A182NAU7 A0A1L8DB20 A0A1B0DG70 A0A1B0CHN4 Q16WP3 A0A1B0GQ66 T1IV58 A0A1Q3F345 B0W9V5 A0A1Q3F2X6 A0A182H047 N6SZB5 A0A0L8HD68 C3YLH1 A0A0K8TQM6 A0A2T7NTN1 A0A336MLY6 A0A087UF34 A0A0P4WJ08 A0A2B4SR23 A0A2C9K2S5 A0A0J7L2U0 A0A182YMP9 V4CBN3 A0A0P4VMH2 A0A0V0G670 T2MAS3 A0A1D2MUG4 A0A210PFP2 A0A0B6ZQH4 K1QH18 A0A224XR85 A7SAR3 Q9VR38 A0A0J9TFN7 B4NZ20 A0A0A1WVC6 A0A2H8TRD8 Q8MRZ0 V5HJG2 B4I174 B4N0J8 A0A034WHA9 B3N4A8 A0A0K2T5W3
Pubmed
19121390
28756777
22118469
26354079
24845553
18362917
+ More
19820115 21347285 21719571 28648823 24508170 20798317 20075255 21282665 30249741 28004739 20920257 23761445 24438588 20966253 12364791 14747013 17210077 23254933 17510324 26483478 23537049 18563158 26369729 15562597 25244985 27129103 24065732 27289101 28812685 22992520 17615350 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17994087 17550304 25830018 25765539 25348373
19820115 21347285 21719571 28648823 24508170 20798317 20075255 21282665 30249741 28004739 20920257 23761445 24438588 20966253 12364791 14747013 17210077 23254933 17510324 26483478 23537049 18563158 26369729 15562597 25244985 27129103 24065732 27289101 28812685 22992520 17615350 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17994087 17550304 25830018 25765539 25348373
EMBL
BABH01018704
BABH01018705
BABH01018706
BABH01018707
KZ150026
PZC74801.1
+ More
NWSH01000053 PCG80171.1 RSAL01000126 RVE46559.1 AGBW02012657 OWR44645.1 ODYU01000435 SOQ35196.1 KQ459588 KPI97750.1 GDQN01000782 JAT90272.1 KQ460417 KPJ14878.1 GDQN01009928 JAT81126.1 NEVH01016358 PNF25223.1 KK852471 KDR23174.1 KQ971352 EFA06262.1 KQ981673 KYN38340.1 ADTU01011283 KQ976500 KYM83208.1 KQ980581 KYN15291.1 GL888932 EGI57213.1 KQ982373 KYQ56986.1 KZ288194 PBC33904.1 NNAY01005719 OXU16578.1 KK107276 EZA53641.1 KQ977242 KYN04681.1 GL447257 EFN86635.1 AAZX01006018 GL762841 EFZ20660.1 GL435766 EFN72720.1 KQ434905 KZC11214.1 KQ414667 KOC64701.1 QOIP01000004 RLU23828.1 PNF25225.1 GEZM01044489 JAV78182.1 GEBQ01025836 JAT14141.1 APCN01001698 ADMH02000283 ETN67125.1 ATLV01021498 KE525319 KFB46609.1 AXCM01000917 AXCN02000656 AAAB01008980 EAA14084.4 GECU01000140 JAT07567.1 AMQN01011686 KB309028 ELT95737.1 GFDF01010544 JAV03540.1 AJVK01059336 AJWK01012538 CH477557 EAT39024.1 AJVK01035595 JH431575 GFDL01013127 JAV21918.1 DS231866 EDS40522.1 GFDL01013142 JAV21903.1 JXUM01100699 KQ564590 KXJ72052.1 APGK01051116 KB741181 KB632333 ENN73099.1 ERL92750.1 KQ418469 KOF87238.1 GG666527 EEN58908.1 GDAI01000954 JAI16649.1 PZQS01000009 PVD24522.1 UFQT01000758 SSX27028.1 KK119542 KFM75973.1 GDRN01054937 JAI66038.1 LSMT01000034 PFX31569.1 LBMM01001064 KMQ96993.1 KB201037 ESO99284.1 GDKW01003085 JAI53510.1 GECL01002556 JAP03568.1 HAAD01002820 CDG69052.1 LJIJ01000508 ODM96727.1 NEDP02076733 OWF35302.1 HACG01024019 CEK70884.1 JH818263 EKC28095.1 GFTR01005905 JAW10521.1 DS469611 EDO39191.1 AE014134 BT150242 AAF50969.1 AGV77144.1 CM002910 KMY88140.1 CM000157 EDW87684.1 GBXI01011949 JAD02343.1 GFXV01004948 MBW16753.1 AY119194 AAM51054.1 GANP01010840 JAB73628.1 CH480820 EDW54281.1 CH963920 EDW77611.1 GAKP01003981 JAC54971.1 CH954177 EDV57778.1 HACA01004052 CDW21413.1
NWSH01000053 PCG80171.1 RSAL01000126 RVE46559.1 AGBW02012657 OWR44645.1 ODYU01000435 SOQ35196.1 KQ459588 KPI97750.1 GDQN01000782 JAT90272.1 KQ460417 KPJ14878.1 GDQN01009928 JAT81126.1 NEVH01016358 PNF25223.1 KK852471 KDR23174.1 KQ971352 EFA06262.1 KQ981673 KYN38340.1 ADTU01011283 KQ976500 KYM83208.1 KQ980581 KYN15291.1 GL888932 EGI57213.1 KQ982373 KYQ56986.1 KZ288194 PBC33904.1 NNAY01005719 OXU16578.1 KK107276 EZA53641.1 KQ977242 KYN04681.1 GL447257 EFN86635.1 AAZX01006018 GL762841 EFZ20660.1 GL435766 EFN72720.1 KQ434905 KZC11214.1 KQ414667 KOC64701.1 QOIP01000004 RLU23828.1 PNF25225.1 GEZM01044489 JAV78182.1 GEBQ01025836 JAT14141.1 APCN01001698 ADMH02000283 ETN67125.1 ATLV01021498 KE525319 KFB46609.1 AXCM01000917 AXCN02000656 AAAB01008980 EAA14084.4 GECU01000140 JAT07567.1 AMQN01011686 KB309028 ELT95737.1 GFDF01010544 JAV03540.1 AJVK01059336 AJWK01012538 CH477557 EAT39024.1 AJVK01035595 JH431575 GFDL01013127 JAV21918.1 DS231866 EDS40522.1 GFDL01013142 JAV21903.1 JXUM01100699 KQ564590 KXJ72052.1 APGK01051116 KB741181 KB632333 ENN73099.1 ERL92750.1 KQ418469 KOF87238.1 GG666527 EEN58908.1 GDAI01000954 JAI16649.1 PZQS01000009 PVD24522.1 UFQT01000758 SSX27028.1 KK119542 KFM75973.1 GDRN01054937 JAI66038.1 LSMT01000034 PFX31569.1 LBMM01001064 KMQ96993.1 KB201037 ESO99284.1 GDKW01003085 JAI53510.1 GECL01002556 JAP03568.1 HAAD01002820 CDG69052.1 LJIJ01000508 ODM96727.1 NEDP02076733 OWF35302.1 HACG01024019 CEK70884.1 JH818263 EKC28095.1 GFTR01005905 JAW10521.1 DS469611 EDO39191.1 AE014134 BT150242 AAF50969.1 AGV77144.1 CM002910 KMY88140.1 CM000157 EDW87684.1 GBXI01011949 JAD02343.1 GFXV01004948 MBW16753.1 AY119194 AAM51054.1 GANP01010840 JAB73628.1 CH480820 EDW54281.1 CH963920 EDW77611.1 GAKP01003981 JAC54971.1 CH954177 EDV57778.1 HACA01004052 CDW21413.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053268
UP000053240
+ More
UP000235965 UP000027135 UP000007266 UP000078541 UP000005205 UP000078540 UP000078492 UP000007755 UP000075809 UP000005203 UP000242457 UP000215335 UP000053097 UP000078542 UP000008237 UP000002358 UP000000311 UP000076502 UP000053825 UP000192223 UP000279307 UP000085678 UP000075840 UP000075881 UP000069272 UP000075900 UP000075903 UP000000673 UP000075902 UP000075885 UP000030765 UP000075920 UP000075883 UP000075886 UP000075882 UP000007062 UP000076407 UP000014760 UP000075880 UP000075884 UP000092462 UP000092461 UP000008820 UP000002320 UP000069940 UP000249989 UP000019118 UP000030742 UP000053454 UP000001554 UP000245119 UP000054359 UP000225706 UP000076420 UP000036403 UP000076408 UP000030746 UP000094527 UP000242188 UP000005408 UP000001593 UP000000803 UP000002282 UP000001292 UP000007798 UP000008711
UP000235965 UP000027135 UP000007266 UP000078541 UP000005205 UP000078540 UP000078492 UP000007755 UP000075809 UP000005203 UP000242457 UP000215335 UP000053097 UP000078542 UP000008237 UP000002358 UP000000311 UP000076502 UP000053825 UP000192223 UP000279307 UP000085678 UP000075840 UP000075881 UP000069272 UP000075900 UP000075903 UP000000673 UP000075902 UP000075885 UP000030765 UP000075920 UP000075883 UP000075886 UP000075882 UP000007062 UP000076407 UP000014760 UP000075880 UP000075884 UP000092462 UP000092461 UP000008820 UP000002320 UP000069940 UP000249989 UP000019118 UP000030742 UP000053454 UP000001554 UP000245119 UP000054359 UP000225706 UP000076420 UP000036403 UP000076408 UP000030746 UP000094527 UP000242188 UP000005408 UP000001593 UP000000803 UP000002282 UP000001292 UP000007798 UP000008711
Pfam
PF03164 Mon1
SUPFAM
SSF64356
SSF64356
ProteinModelPortal
H9JWU3
A0A2W1BKJ9
A0A2A4K7G1
A0A3S2NRA8
A0A212ET21
A0A2H1V351
+ More
A0A194Q2W4 A0A1E1WTH9 A0A0N0PDA4 A0A1E1W2B9 A0A2J7Q9G0 A0A067RTL3 D6WU67 A0A195FCH7 A0A158NBQ7 A0A195BF61 A0A195DR18 F4X862 A0A151X9C3 A0A087ZZR2 A0A2A3EQX7 A0A232EE22 A0A026WDC2 A0A151IKJ4 E2BC99 K7IUN0 E9IFJ1 E2A1P7 A0A154PI96 A0A0L7R1M9 A0A1W4WY10 A0A3L8DVP5 A0A2J7Q9G6 A0A1Y1LXA3 A0A1B6KRU6 A0A1S3H631 A0A182HRF5 A0A182KAU8 A0A182F1G2 A0A182RRA7 A0A182UTG5 W5JVP3 A0A182TUG4 A0A182PG39 A0A084W8R5 A0A182W8Y3 A0A182MTV1 A0A182QTR3 A0A182KYX7 Q7Q176 A0A182X2I9 A0A1B6K7Z4 R7TQ94 A0A182J4D9 A0A182NAU7 A0A1L8DB20 A0A1B0DG70 A0A1B0CHN4 Q16WP3 A0A1B0GQ66 T1IV58 A0A1Q3F345 B0W9V5 A0A1Q3F2X6 A0A182H047 N6SZB5 A0A0L8HD68 C3YLH1 A0A0K8TQM6 A0A2T7NTN1 A0A336MLY6 A0A087UF34 A0A0P4WJ08 A0A2B4SR23 A0A2C9K2S5 A0A0J7L2U0 A0A182YMP9 V4CBN3 A0A0P4VMH2 A0A0V0G670 T2MAS3 A0A1D2MUG4 A0A210PFP2 A0A0B6ZQH4 K1QH18 A0A224XR85 A7SAR3 Q9VR38 A0A0J9TFN7 B4NZ20 A0A0A1WVC6 A0A2H8TRD8 Q8MRZ0 V5HJG2 B4I174 B4N0J8 A0A034WHA9 B3N4A8 A0A0K2T5W3
A0A194Q2W4 A0A1E1WTH9 A0A0N0PDA4 A0A1E1W2B9 A0A2J7Q9G0 A0A067RTL3 D6WU67 A0A195FCH7 A0A158NBQ7 A0A195BF61 A0A195DR18 F4X862 A0A151X9C3 A0A087ZZR2 A0A2A3EQX7 A0A232EE22 A0A026WDC2 A0A151IKJ4 E2BC99 K7IUN0 E9IFJ1 E2A1P7 A0A154PI96 A0A0L7R1M9 A0A1W4WY10 A0A3L8DVP5 A0A2J7Q9G6 A0A1Y1LXA3 A0A1B6KRU6 A0A1S3H631 A0A182HRF5 A0A182KAU8 A0A182F1G2 A0A182RRA7 A0A182UTG5 W5JVP3 A0A182TUG4 A0A182PG39 A0A084W8R5 A0A182W8Y3 A0A182MTV1 A0A182QTR3 A0A182KYX7 Q7Q176 A0A182X2I9 A0A1B6K7Z4 R7TQ94 A0A182J4D9 A0A182NAU7 A0A1L8DB20 A0A1B0DG70 A0A1B0CHN4 Q16WP3 A0A1B0GQ66 T1IV58 A0A1Q3F345 B0W9V5 A0A1Q3F2X6 A0A182H047 N6SZB5 A0A0L8HD68 C3YLH1 A0A0K8TQM6 A0A2T7NTN1 A0A336MLY6 A0A087UF34 A0A0P4WJ08 A0A2B4SR23 A0A2C9K2S5 A0A0J7L2U0 A0A182YMP9 V4CBN3 A0A0P4VMH2 A0A0V0G670 T2MAS3 A0A1D2MUG4 A0A210PFP2 A0A0B6ZQH4 K1QH18 A0A224XR85 A7SAR3 Q9VR38 A0A0J9TFN7 B4NZ20 A0A0A1WVC6 A0A2H8TRD8 Q8MRZ0 V5HJG2 B4I174 B4N0J8 A0A034WHA9 B3N4A8 A0A0K2T5W3
PDB
5LDD
E-value=1.04553e-18,
Score=231
Ontologies
KEGG
GO
PANTHER
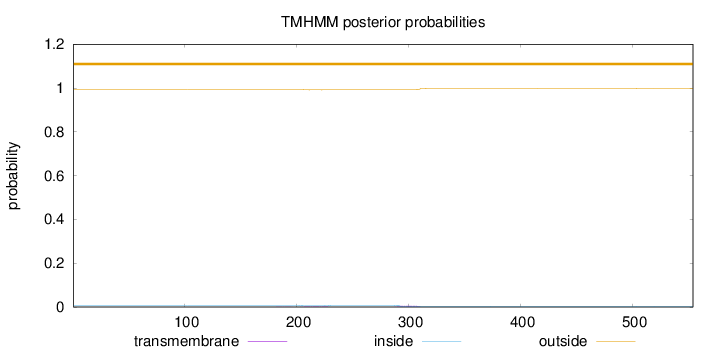
Topology
Length:
554
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15748
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00677
outside
1 - 554
Population Genetic Test Statistics
Pi
253.791031
Theta
189.453122
Tajima's D
1.528925
CLR
0.327728
CSRT
0.791460426978651
Interpretation
Uncertain
