Gene
KWMTBOMO16284
Pre Gene Modal
BGIBMGA014005
Annotation
PREDICTED:_neuronal_acetylcholine_receptor_subunit_beta-3-like?_partial_[Papilio_xuthus]
Full name
Mitochondrial genome maintenance exonuclease 1
Location in the cell
PlasmaMembrane Reliability : 3.565
Sequence
CDS
ATGAAACGGACTGTGTTCATAATCTGGTTACTCGCATGTGTGTGTACAGTTAGAATGGATAATTTCACTTATAGTTACAAAGATGCGATGCGCTGTAAAGAGCATATCTGTAGACACGGTTACAGTTACGAAACTTTCTACCGGGTGGACGCGAACGAAATGAGGAGCATCAACGTTGACCCTCATTCAGTCTTCGAGATGCACATCGCGATCCAAGCCGCCAGCAACGGACACATAGTGCTCTCTGAAATCCCTAATCCTGGCGTCAGTGATCCAGTTTACGAAATTGTGGTCGGCGGTGGAGGGAACAAGTTCACCGAACTACGCCGCAACCTGCGACGCAATGCCAAGAGCTCCGTCACCACACCCCAAATATTGTCCCCTATAGAACTCAGAGGCTTCTACATCAAAATATCTGAAGATGGCCTGGTAGAATTCGGTAGAGAAGGACAGGCACTACCTTTGATAAGCTTCAATGATGTCGACCCATTATCCGTGAAGTACTTCAGCTTCGCGGCCTGGAACGGCGTTGAAGCCAAATTCTTGTACGACTGCCCTATACCCGGAGACACGTCTATTGAAGACATCCCTAACTCCCACCCAGAAGAGCCCAAGCTAACTCCTACGGACAAGTTAAAGCGGACTTTGCTAATGGGTCGGCTTCCCTCTGTCCCGCCCAATCCCGTCATGGAAGTAAAACTCGGATTTAAACTAACCAGCTTTCGGTATAACACTTTGGACTCCAAATTGACGACCACAGCGGCTGTTGTCGTGTCATGGGAAGATGATAGCATGGCGTGGAATCCGGACAAATTCAACGGAACCACCAACATCAAATTACGACAAGGCCAGATTTGGAAACCCTCGTTCTACGTTTTAAATTCGGATGGATATGGCGCGATGGATTCAACGAATCCGGATACAATTTCGTTGGCCAGCAGCGGGGAGGCGACATTGCACTTTCAGACCCAAACGCAGAACTATTGCTTCGATACAACAACGACTTACAGCAGATGGCCGCATGATGTATACCAGTGTCTGATCGTCATAGAGCCTTGGGAGGCCCACGAAAACATAAAAATTGAAATACTCGACGAGACTGATGCGAAGTTCGAAATGTTTTCAGATATTGACACTGTCATCCATAACGAATGGCAAATCATGACAGAGCAAGTCACTCTTGATTCAGCGATGTGGAACGAGATCATCGACCTCATCAACAGCGAGGAGGTTCTGAACACCCATTTAAGTGACAAGATGATCATCACTGTGGACTTAGCCAAAAACCCTGACTTGTACAACCTGGTGTTCTACGTGCCTATGTTGGTCCTAAACACGTTCGTCTTAATATCGTTCTGGACGGAGCCACTGACAATGTCAAGGATATGGTTCTACGTAGCCTGTGTGGTGTCTATCTGTTTTGGACTCTGCTATATCGATTATTTAATACCGATTCATGCGATACCTACCATAATGGTGCTGTACGTGACGGTCTTGTTTGGACTAATGACTGCTGTGATGGCCAACGTGCTCCTAATATCGAACTTCGGTGAGGCGTTGGGGAAGTCGTCAGCAATGAAACAAGTCCTGACCTGCGGCGTTGTACGAGCGGTCTTCTGTTTGCCTTCCATGAAGAAAGCCGCCAATGGGGATTACTCGGTCCAAGAAGATGAGGACTCTAGTTACATAGTGGATTCGCGGGAAGGCGTTGAAGAGATGGAGAGGAGTGACAAAGAAATAAAATACGGTGACAAGGCCGAACTAGCGGTGGTCGTTGATAAAATCCTTTTCGTTGTCTACCTTATTGTTTTTGTCGGAATGTTGGCAGCGCATTACTGA
Protein
MKRTVFIIWLLACVCTVRMDNFTYSYKDAMRCKEHICRHGYSYETFYRVDANEMRSINVDPHSVFEMHIAIQAASNGHIVLSEIPNPGVSDPVYEIVVGGGGNKFTELRRNLRRNAKSSVTTPQILSPIELRGFYIKISEDGLVEFGREGQALPLISFNDVDPLSVKYFSFAAWNGVEAKFLYDCPIPGDTSIEDIPNSHPEEPKLTPTDKLKRTLLMGRLPSVPPNPVMEVKLGFKLTSFRYNTLDSKLTTTAAVVVSWEDDSMAWNPDKFNGTTNIKLRQGQIWKPSFYVLNSDGYGAMDSTNPDTISLASSGEATLHFQTQTQNYCFDTTTTYSRWPHDVYQCLIVIEPWEAHENIKIEILDETDAKFEMFSDIDTVIHNEWQIMTEQVTLDSAMWNEIIDLINSEEVLNTHLSDKMIITVDLAKNPDLYNLVFYVPMLVLNTFVLISFWTEPLTMSRIWFYVACVVSICFGLCYIDYLIPIHAIPTIMVLYVTVLFGLMTAVMANVLLISNFGEALGKSSAMKQVLTCGVVRAVFCLPSMKKAANGDYSVQEDEDSSYIVDSREGVEEMERSDKEIKYGDKAELAVVVDKILFVVYLIVFVGMLAAHY
Summary
Description
Metal-dependent single-stranded DNA (ssDNA) exonuclease involved in mitochondrial genome maintenance.
Similarity
Belongs to the glycosyl hydrolase 18 family.
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Belongs to the MGME1 family.
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Belongs to the MGME1 family.
Uniprot
A0A2A4J1J6
A0A194PWK2
A0A2W1BIB0
A0A3S2PBC3
A0A0N1I8C1
A0A139WFG7
+ More
A0A139WFS6 A0A1Y1KLR1 A0A2P8YKE5 A0A1Q3FMQ8 A0A1B6EI03 N6T616 A0A1B6FWE9 B0WG41 A0A023EXS4 A0A182GUM0 A0A0P4VUH2 A0A1W5YMS4 A0A0P6K120 J9JMD6 A0A0N8AQU7 A0A0P5GAT8 A0A0P5ADJ4 A0A1S4F984 Q17BJ5 A0A0P6I592 A0A2Z4N5W6 A0A146LI87 A0A0M4EIV7 A0A0N8DLD4 B4P8A1 A0A0M3QVI3 B4LNF1 A0A1I8MXG6 A0A0L0CQE0 A0A0M5J6Q8 B3NND4 A0A224XKR8 B4I805 A0A1W4V0S7 A0A1A9YKU4 A0A0A1X6Z4 A0A146KW52 H8F4S1 Q9W288 B4KMU7 A0A0B4K8A6 B4MNS9 A0A182JN83 B4QH26 A0A1A9V5D9 B4H6X9 A0A1I8PIZ3 E0VC57 B3MBI7 Q28WM6 A0A1A9WIU0 A0A034W0B7 B4J8I8 A0A182KD66 A0A182P8G7 A0A1B0G7M4 A0A1S4H4F3 A0A3B0J1L1 A0A182HFQ8 A0A182X1U3 A0A182MYE1 A0A1A9ZGK1 A0A0K8ULU6
A0A139WFS6 A0A1Y1KLR1 A0A2P8YKE5 A0A1Q3FMQ8 A0A1B6EI03 N6T616 A0A1B6FWE9 B0WG41 A0A023EXS4 A0A182GUM0 A0A0P4VUH2 A0A1W5YMS4 A0A0P6K120 J9JMD6 A0A0N8AQU7 A0A0P5GAT8 A0A0P5ADJ4 A0A1S4F984 Q17BJ5 A0A0P6I592 A0A2Z4N5W6 A0A146LI87 A0A0M4EIV7 A0A0N8DLD4 B4P8A1 A0A0M3QVI3 B4LNF1 A0A1I8MXG6 A0A0L0CQE0 A0A0M5J6Q8 B3NND4 A0A224XKR8 B4I805 A0A1W4V0S7 A0A1A9YKU4 A0A0A1X6Z4 A0A146KW52 H8F4S1 Q9W288 B4KMU7 A0A0B4K8A6 B4MNS9 A0A182JN83 B4QH26 A0A1A9V5D9 B4H6X9 A0A1I8PIZ3 E0VC57 B3MBI7 Q28WM6 A0A1A9WIU0 A0A034W0B7 B4J8I8 A0A182KD66 A0A182P8G7 A0A1B0G7M4 A0A1S4H4F3 A0A3B0J1L1 A0A182HFQ8 A0A182X1U3 A0A182MYE1 A0A1A9ZGK1 A0A0K8ULU6
EC Number
3.1.-.-
Pubmed
EMBL
NWSH01003745
PCG65861.1
KQ459588
KPI97751.1
KZ150026
PZC74799.1
+ More
RSAL01000126 RVE46553.1 KQ460417 KPJ14879.1 KQ971352 KYB26738.1 KYB26739.1 GEZM01084981 JAV60346.1 PYGN01000530 PSN44731.1 GFDL01006210 JAV28835.1 GECZ01032221 JAS37548.1 APGK01051097 APGK01051098 APGK01051099 APGK01051100 KB741181 ENN73093.1 GECZ01015300 JAS54469.1 DS231923 EDS26737.1 GBBI01004921 JAC13791.1 JXUM01089653 JXUM01089654 JXUM01089655 JXUM01089656 KQ563786 KXJ73306.1 GDKW01002108 JAI54487.1 KX426067 ARI45492.1 GDUN01000307 JAN95612.1 ABLF02036863 GDIQ01251883 JAJ99841.1 GDIQ01248929 JAK02796.1 GDIP01213390 JAJ10012.1 CH477321 EAT43615.1 GDIQ01009267 JAN85470.1 MF612141 AWX65629.1 GDHC01012449 JAQ06180.1 CP012524 ALC42797.1 GDIP01022375 JAM81340.1 CM000158 EDW91141.1 ALC42433.1 CH940648 EDW61103.1 JRES01000171 KNC33654.1 ALC40474.1 CH954179 EDV55558.1 GFTR01006008 JAW10418.1 CH480824 EDW56730.1 GBXI01007864 JAD06428.1 GDHC01018764 JAP99864.1 BT133311 AFD10758.1 AE013599 AAF46805.1 CH933808 EDW09869.2 AFH08210.1 CH963848 EDW73768.2 CM000362 CM002911 EDX08172.1 KMY95727.1 CH479216 EDW33615.1 DS235047 EEB10963.1 CH902619 EDV37118.1 CM000071 EAL26641.2 GAKP01011352 JAC47600.1 CH916367 EDW01255.1 CCAG010016887 AAAB01008933 OUUW01000001 SPP72752.1 APCN01005565 GDHF01024788 JAI27526.1
RSAL01000126 RVE46553.1 KQ460417 KPJ14879.1 KQ971352 KYB26738.1 KYB26739.1 GEZM01084981 JAV60346.1 PYGN01000530 PSN44731.1 GFDL01006210 JAV28835.1 GECZ01032221 JAS37548.1 APGK01051097 APGK01051098 APGK01051099 APGK01051100 KB741181 ENN73093.1 GECZ01015300 JAS54469.1 DS231923 EDS26737.1 GBBI01004921 JAC13791.1 JXUM01089653 JXUM01089654 JXUM01089655 JXUM01089656 KQ563786 KXJ73306.1 GDKW01002108 JAI54487.1 KX426067 ARI45492.1 GDUN01000307 JAN95612.1 ABLF02036863 GDIQ01251883 JAJ99841.1 GDIQ01248929 JAK02796.1 GDIP01213390 JAJ10012.1 CH477321 EAT43615.1 GDIQ01009267 JAN85470.1 MF612141 AWX65629.1 GDHC01012449 JAQ06180.1 CP012524 ALC42797.1 GDIP01022375 JAM81340.1 CM000158 EDW91141.1 ALC42433.1 CH940648 EDW61103.1 JRES01000171 KNC33654.1 ALC40474.1 CH954179 EDV55558.1 GFTR01006008 JAW10418.1 CH480824 EDW56730.1 GBXI01007864 JAD06428.1 GDHC01018764 JAP99864.1 BT133311 AFD10758.1 AE013599 AAF46805.1 CH933808 EDW09869.2 AFH08210.1 CH963848 EDW73768.2 CM000362 CM002911 EDX08172.1 KMY95727.1 CH479216 EDW33615.1 DS235047 EEB10963.1 CH902619 EDV37118.1 CM000071 EAL26641.2 GAKP01011352 JAC47600.1 CH916367 EDW01255.1 CCAG010016887 AAAB01008933 OUUW01000001 SPP72752.1 APCN01005565 GDHF01024788 JAI27526.1
Proteomes
UP000218220
UP000053268
UP000283053
UP000053240
UP000007266
UP000245037
+ More
UP000019118 UP000002320 UP000069940 UP000249989 UP000007819 UP000008820 UP000092553 UP000002282 UP000008792 UP000095301 UP000037069 UP000008711 UP000001292 UP000192221 UP000092443 UP000000803 UP000009192 UP000007798 UP000075880 UP000000304 UP000078200 UP000008744 UP000095300 UP000009046 UP000007801 UP000001819 UP000091820 UP000001070 UP000075881 UP000075885 UP000092444 UP000268350 UP000075840 UP000076407 UP000075884 UP000092445
UP000019118 UP000002320 UP000069940 UP000249989 UP000007819 UP000008820 UP000092553 UP000002282 UP000008792 UP000095301 UP000037069 UP000008711 UP000001292 UP000192221 UP000092443 UP000000803 UP000009192 UP000007798 UP000075880 UP000000304 UP000078200 UP000008744 UP000095300 UP000009046 UP000007801 UP000001819 UP000091820 UP000001070 UP000075881 UP000075885 UP000092444 UP000268350 UP000075840 UP000076407 UP000075884 UP000092445
Pfam
Interpro
IPR006201
Neur_channel
+ More
IPR022041 Methyltransf_FA
IPR036734 Neur_chan_lig-bd_sf
IPR006202 Neur_chan_lig-bd
IPR017853 Glycoside_hydrolase_SF
IPR036508 Chitin-bd_dom_sf
IPR011583 Chitinase_II
IPR029070 Chitinase_insertion_sf
IPR001223 Glyco_hydro18_cat
IPR001579 Glyco_hydro_18_chit_AS
IPR002557 Chitin-bd_dom
IPR036719 Neuro-gated_channel_TM_sf
IPR006029 Neurotrans-gated_channel_TM
IPR038726 PDDEXK_AddAB-type
IPR019406 APLF_PBZ
IPR026970 MGME1
IPR027415 TDP_C
IPR010347 Tdp1
IPR022041 Methyltransf_FA
IPR036734 Neur_chan_lig-bd_sf
IPR006202 Neur_chan_lig-bd
IPR017853 Glycoside_hydrolase_SF
IPR036508 Chitin-bd_dom_sf
IPR011583 Chitinase_II
IPR029070 Chitinase_insertion_sf
IPR001223 Glyco_hydro18_cat
IPR001579 Glyco_hydro_18_chit_AS
IPR002557 Chitin-bd_dom
IPR036719 Neuro-gated_channel_TM_sf
IPR006029 Neurotrans-gated_channel_TM
IPR038726 PDDEXK_AddAB-type
IPR019406 APLF_PBZ
IPR026970 MGME1
IPR027415 TDP_C
IPR010347 Tdp1
SUPFAM
Gene 3D
ProteinModelPortal
A0A2A4J1J6
A0A194PWK2
A0A2W1BIB0
A0A3S2PBC3
A0A0N1I8C1
A0A139WFG7
+ More
A0A139WFS6 A0A1Y1KLR1 A0A2P8YKE5 A0A1Q3FMQ8 A0A1B6EI03 N6T616 A0A1B6FWE9 B0WG41 A0A023EXS4 A0A182GUM0 A0A0P4VUH2 A0A1W5YMS4 A0A0P6K120 J9JMD6 A0A0N8AQU7 A0A0P5GAT8 A0A0P5ADJ4 A0A1S4F984 Q17BJ5 A0A0P6I592 A0A2Z4N5W6 A0A146LI87 A0A0M4EIV7 A0A0N8DLD4 B4P8A1 A0A0M3QVI3 B4LNF1 A0A1I8MXG6 A0A0L0CQE0 A0A0M5J6Q8 B3NND4 A0A224XKR8 B4I805 A0A1W4V0S7 A0A1A9YKU4 A0A0A1X6Z4 A0A146KW52 H8F4S1 Q9W288 B4KMU7 A0A0B4K8A6 B4MNS9 A0A182JN83 B4QH26 A0A1A9V5D9 B4H6X9 A0A1I8PIZ3 E0VC57 B3MBI7 Q28WM6 A0A1A9WIU0 A0A034W0B7 B4J8I8 A0A182KD66 A0A182P8G7 A0A1B0G7M4 A0A1S4H4F3 A0A3B0J1L1 A0A182HFQ8 A0A182X1U3 A0A182MYE1 A0A1A9ZGK1 A0A0K8ULU6
A0A139WFS6 A0A1Y1KLR1 A0A2P8YKE5 A0A1Q3FMQ8 A0A1B6EI03 N6T616 A0A1B6FWE9 B0WG41 A0A023EXS4 A0A182GUM0 A0A0P4VUH2 A0A1W5YMS4 A0A0P6K120 J9JMD6 A0A0N8AQU7 A0A0P5GAT8 A0A0P5ADJ4 A0A1S4F984 Q17BJ5 A0A0P6I592 A0A2Z4N5W6 A0A146LI87 A0A0M4EIV7 A0A0N8DLD4 B4P8A1 A0A0M3QVI3 B4LNF1 A0A1I8MXG6 A0A0L0CQE0 A0A0M5J6Q8 B3NND4 A0A224XKR8 B4I805 A0A1W4V0S7 A0A1A9YKU4 A0A0A1X6Z4 A0A146KW52 H8F4S1 Q9W288 B4KMU7 A0A0B4K8A6 B4MNS9 A0A182JN83 B4QH26 A0A1A9V5D9 B4H6X9 A0A1I8PIZ3 E0VC57 B3MBI7 Q28WM6 A0A1A9WIU0 A0A034W0B7 B4J8I8 A0A182KD66 A0A182P8G7 A0A1B0G7M4 A0A1S4H4F3 A0A3B0J1L1 A0A182HFQ8 A0A182X1U3 A0A182MYE1 A0A1A9ZGK1 A0A0K8ULU6
PDB
4PIR
E-value=8.42924e-05,
Score=111
Ontologies
GO
Topology
Subcellular location
Mitochondrion
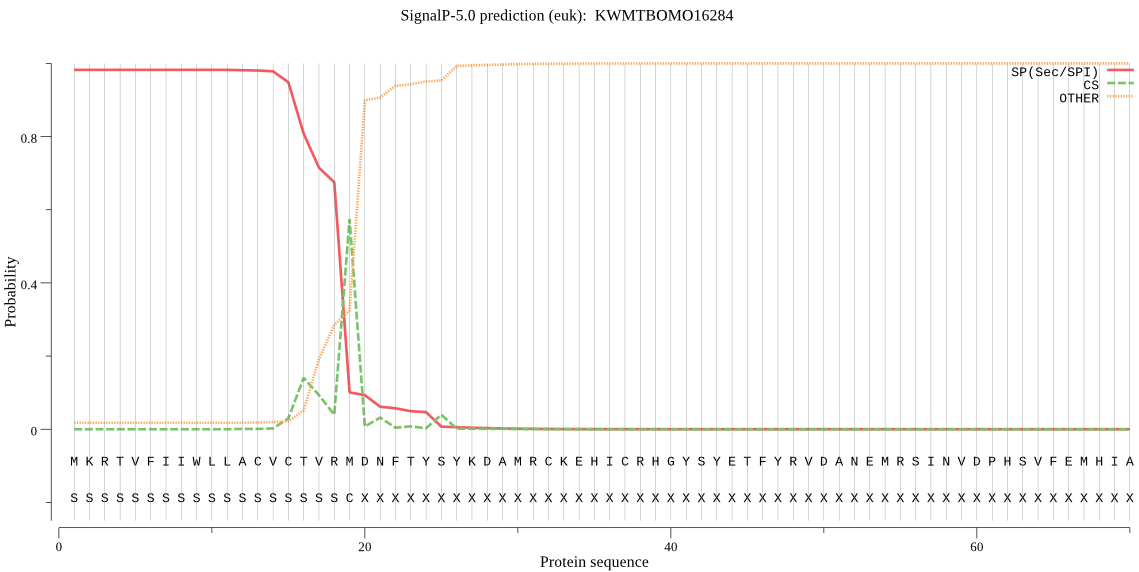
SignalP
Position: 1 - 19,
Likelihood: 0.981931
Length:
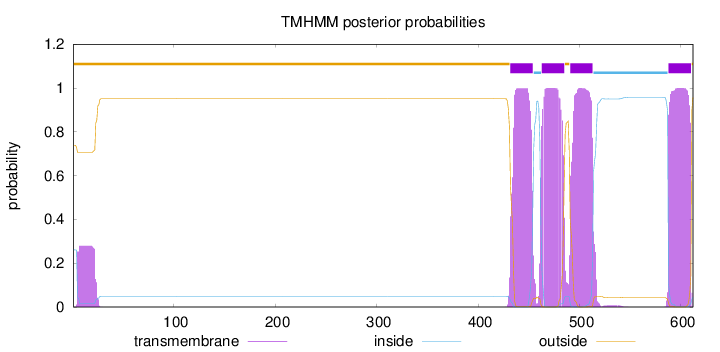
612
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
94.64836
Exp number, first 60 AAs:
5.27883
Total prob of N-in:
0.26125
outside
1 - 431
TMhelix
432 - 454
inside
455 - 462
TMhelix
463 - 485
outside
486 - 490
TMhelix
491 - 513
inside
514 - 587
TMhelix
588 - 610
outside
611 - 612
Population Genetic Test Statistics
Pi
165.759904
Theta
150.419812
Tajima's D
0.21695
CLR
236.630448
CSRT
0.429778511074446
Interpretation
Uncertain
