Gene
KWMTBOMO16269 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013894
Annotation
uncharacterized_LOC101743290_precursor_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.158 PlasmaMembrane Reliability : 1.384
Sequence
CDS
ATGAACAAACGCATCAGCGTGGTGCTGAACGTCATACTTCCGAAGCCAAACGTGGATGTGCACACGCTGTGCGTCGACGGCGACCTGGGCGGCGACCAGAGCCCGATCAAGACTATAGACGCCAAGTACAAGACTGACGTCACCAAGATCCTGACCGCCATAAAAGGAAACCTGATACAACTACCGGAAAGCTTCGAATCGAATTCGACTGTGACTTGGACGTCGGGCGCCGAGTACAAGAACATAGAGAACTACATAGACCACAAGTGGGACATTAACGGTTCCCACTTCATAGACTACACTCTAGTGACGCCGCTCTACGAGAACGACTCTACGTTCAAGGTCAAGGGATCGTACCAGAGGGATTTGGTGCACAAATTGGAGATTGTCAAAGGCCACATGCACAAGCCTGGTGCATCCCAGATCGGCGAGATGGACGTGCGTTACGGCGGCGTGAAACACACAGACGGATACTTCAACTTCACGACGCCCTTCCAGAAACTGAAATGGCTCAAATCTATATTTGACATCAACAATATTGAGGAGCATTCCGACAACAAAGTGAACTTGTATTGGCCGAACAAGACAGCGTCCGTGAACACGACCCACCAGTACTCGAAGGACGCGGGCGGGTTCAAGCAGACCGGACAAATAGCTCTGTCCGTTCCGCTCAGCACCCAGCACTTGGTCAACACCGAGTATTATTATCTGCAAGGAGACAAGTCTAGTAACGGTAACGCGACCGTCGATCTGGACGGGGATCGGTTCGTGAAGGGCTCCTTCAAACAAATCCTCAGTAAAAGCGAAAGGAATTTGGACCTTGCCACGACGGACATAGAGGTCGAAAACGTACACACGCCGGTCGGCGTTAAGTACATACACGAATACGACGAAACCGGGAAAGTTGACGTGAAGCAAGCGACCGTGTTCCACCTGCACAACGCGACCAAGTTCAACGTGACCGGGAAGCTGGACGTGCTCACGTACGACACGGGCAAGGACCTGAGGCTCACCGCCGTCCACGGCACCCGCACCTGGACCTTCGACAACCACTACGAGGCCTTCGACAAGGAGCTCAAGCAGGGCAGCAAGATTAAATGGATCGAAGACTACTGGATCAACTATGACATCCACGTCACTAATATGACGACGGCGGACACCGAATCGCAGCAACTCGTGATGAATGTTTACTATCCATTGCGGACCTTCAATCTGGTAGCGGTATACAACTTGCAAGACACCCTGCTCGATGGTAAAGCACAGCTGAACTGGGACGTGAAAGCCGAGAACAAAACAGCCGAATTGAGAGGCAGGTGGGAGAACCCCCCCATAAAAGAGGGGAACATGCATAACATCGATTTGATACTATCGCATCCTTCGTTCAGGAAGGACGTGAACTTGAAAGCTCAGTACTCGAGCACGCCAGCGGTGATGTCGAACGTGTCGCTAGAGCTGCAATACTCGGACATAGAGAGCGAATATCTAAAATTAAAATCGATTCTAACCGATAACTCGAATGGTCCGATACGCGATTACTCATTCGCGCTGACCTGCACGCACCCCTCCACGTACCTAGACCTGGACATGAAGTCCGACGTGAACATACACAGCAAGTGGTATTACTTCTACAACTACTACAGGTTCCAGAAGTCGTTGTATTACGAGAAGATAAGGCAGAGCAAGCTCGTGGTGGACACCACGAAGAGCTTCGTCAATTGGGAGCGCACCAACGAGACGTACTTCTACAGAGTGAACGGCACTTGGGACCTCACGTATCCGGACTACACGATAAAGTCGTTCATACATCGCACCACCGGCAACGACACCGGTTTGGCCGTGCTCTCAATGAAAGACAAGTCCTTGGTCGCCCATCTGAATAGTACTGACGATATATCGTACCATCTAGTTGGTAACATTGTAAACACGAGATCAGCGAAACTAGACGCGTGGCGCGACTTCGATGACGTCACAACCGTAGACCTGGCTTCGTACATCCGCCTCAACCACTCCCGACTTTTGACCAGCTCGCTGGTGTGGCGGCCGCAGATCTTCAGCGAAGTCAAAGCTCAAGCGGTCTCCACGTTGAAGCTTTTATACGGCCAAATAAACGACAGTCTGGTCATACTGAAAGAAGCGCCCATGGAAGCTCACTTGGCGCTCAAAGCCATATGGGCAGACGCCAGACCTAGGATCGGTGATTTCTTGCACGACTTAGACGACCTGCAAGTGATCAAAGACGATTTAGACGACTTTGAAACATTCTTAAACGAATCCTACAGTAAAAATGATTTTTACGTGAAAGACATAGTCGAGTTTACTTACTACGTGCTAGACGAGATGGCCATAAGGAATCACTTGGAGAGTCTGCCCGGCATAGTTAACGACATGTGGGGGATGATGGGCAACACCAGTCAGTCCATCAAGCAGAGTCTGACATACGTGGTCGACACTATAAAACACGCCTACGCGAATTTCTTGGAGTCCGTGAACAAGGCTCTGGAGGCAGATTTTATGGAACTGGTGTCGGAGCGACTCGAAGCCATGATATTACAGTACGATAACTTCATCAGGGACCTCCACATGAAATTGTTGGAGTATTGGGAGGAGACTTGGGTGAACGCGACCAATCGTTTGACCAAATATTGGCACGAACTGCTAAAGTCTATTGAGCCGCTGTTCTTCAAGATCCTGCATTACAGCGAGTCGTTCGTTTTCACGGTGTGGAAGAGTCTGATGGACTTCTTTTACAATCGCACCCAGGAATTGACGGATTCGCCGTATTTCAACTACGTTTCGACGTTTGGTCATGAAATGGACAAAATCTACAAGGACCTGGTCAATAACGACTTGATAACGAACATAAAGAAGTACAGCAAGAAACTGTGGGATGCTGTCTGGACCAAGATAGAGAAATACATCCCGTTCAAAGACGAGATAGTGCAGCTTTACAACGAATTCAAGGACAGCTGGCAGGACTTCCTCAAGACCCCGCAGGTCGTCTACGTCAGGGAGAAGTACCAAGAAGCCTACGTCCGTCTGAAGTGGTGGTACGACTACTTCCTGATCGGCGAGGCCTTGAACACGATCGGCGAAATCATCTACGTGAAGCTCACGGACTTGGCGAAGACTGCACTGCAGTACGAAGAACTGCACCGCACGCCCAAAACCAACTTCATATTCGATCCTCGAGTGGGGGAGATCGTCCTCGAGCAGAAGCTGCCGATGTCCTGGCACGCCTTCAACAGTTCGCCGGACTTCACTGAGATATCCGAGTACAAGACCGTCCGCGACTTTTTCGACGAGTGGCTCACCACCAACAAGAGCATCTGGTCCTACTACTACGAGATAAGGCCGTACATGGACCTCAACAACTTGATGCCGCCTTTCCCTGGCATAGCAATGATGACGGAACAAGGCGCCCTGGTCACTTACGACAAGCAAGTGTTCACGGTGTCCGAGCGCGGCGTGTTCCTGCTCACGAAGGACTACAGGCGCGACAACTTCACCGTGCTCATGGCTTCCAACGACAGAGGCACATACGACCTCGTGCTCGTGACCAGAACGAAACTGGTCTACATTGATCTTTCTAAAGAGCAAGTGACGGTGAGCGACACGGTGCTGAGTCTGCCGGCCTCGGTCGAGGGTCTGGTGATAGACCGGCAGATGGAGACCCTGAGCGTGCGCGGAGACAGCGGCCTCGACGTGCAGTGCAACCTGCGCTTCAAGTCCTGCAAGCTGGGTGTCGAAGGTTGGTTCTACGCGACCCTGGGTGGGTTGCTGGGAACGTACAACAACGAGCAGTACGACGACCTGCAACTGCCGAACAACAGCTTCGCGACCAGCGTCGGCGCGCTGGCGACGGCGTGGCGCCTCGGGCCCGCGCCCCCCGCCCCCGCGCCGCTCCCCGCACCTCCCACCCCGCACAACGAGACGCAGTGCGAGAAGTTCTTCCAGAATAAAGTTTCGCCCTTCCATCCCTGTTTCTCTGTGATCAAAGCGGCGCCGTTCTACTCGGAGTGCCAGGCGGGCGTGGAGGCGTGCGCGCTCGCCAGTGCGTACCTGGAACTGTGCACGCAGCAACACGTGCCCACGCACATGCCGGACCACTGCAACCAGTGCACGACCCCGCACGGTGACGTCATAGACGAAGGGTCCTTCTACACGCTGGAAAACGTGCCAGAATCGACGGACGTGGTGTTCATAATCGAAGCGCAGGTCTGCAACAAGAACGTTAGGAAAACGAAAAATCTCGATCTCTTCCTGGAGACCTTCGACATACAACTGCAGGCCGGCGGCCTCACCGACAACCGGTACGCGGTGGTCGGGTTCGGCGGGCGCGGCGTGTACAGACAGCCGAGGGCGGTGTACGTCAACAACAAAGTGTTCACTGACCCAATAGAAATATCGTCGCACTTCGACACTTTCCAAATCGATAAGTTAGATTTGGCGCGCCCTAACAAGACCGTCAAGGCGGATATGTTCAGCGCTCTGAACTTCGCAACAAAGCTGCCGTTCCGGCCCGGCGTTCCCCGCACCTTCCTGCTGATGCCCTGCACCAGATGCGACTCGCAGTTCATGAGGTTGGACTACTCATCGATGTATCAAAGTTTGATGGAGAATTCAGTAACGCTTCATATTTTAATGGATGACGACTTCACACTATCGAAGAAAAGGGCATCCAAATATTTGTTCGGTGTGGACAGCACGCTGGCTTATACGAACAAGGATTACGAGAAGCTGGTCGGAGACGCAGCTCTCAGGAAACAAGTCAAATTGCCGAAAGATAAACTGGGACTGTGCATCTCCCTTGCCATGGAAACGAATGGTACGATCCTGGCGGGCACGAAGCTGAGTGCGGAGCGTCGCTCGGCGCGGCGGTTCTCGTCCGTGGCGGGCGCGCGGGCGGCGCGCGGGGCGGCGCGGTGCGCGCGCACACGCTGCGAGTGCCGCGCCGGCGCCACGCACTGCCGCCCCTGCGCCGACACACTCAGAATGTGGGAATTAGCATTCTTCAGCACAGACGACATTGACGACTTAATTGACATGGCGATGGAGCCCCCCACCATACCGTCGCTTAGTTGA
Protein
MNKRISVVLNVILPKPNVDVHTLCVDGDLGGDQSPIKTIDAKYKTDVTKILTAIKGNLIQLPESFESNSTVTWTSGAEYKNIENYIDHKWDINGSHFIDYTLVTPLYENDSTFKVKGSYQRDLVHKLEIVKGHMHKPGASQIGEMDVRYGGVKHTDGYFNFTTPFQKLKWLKSIFDINNIEEHSDNKVNLYWPNKTASVNTTHQYSKDAGGFKQTGQIALSVPLSTQHLVNTEYYYLQGDKSSNGNATVDLDGDRFVKGSFKQILSKSERNLDLATTDIEVENVHTPVGVKYIHEYDETGKVDVKQATVFHLHNATKFNVTGKLDVLTYDTGKDLRLTAVHGTRTWTFDNHYEAFDKELKQGSKIKWIEDYWINYDIHVTNMTTADTESQQLVMNVYYPLRTFNLVAVYNLQDTLLDGKAQLNWDVKAENKTAELRGRWENPPIKEGNMHNIDLILSHPSFRKDVNLKAQYSSTPAVMSNVSLELQYSDIESEYLKLKSILTDNSNGPIRDYSFALTCTHPSTYLDLDMKSDVNIHSKWYYFYNYYRFQKSLYYEKIRQSKLVVDTTKSFVNWERTNETYFYRVNGTWDLTYPDYTIKSFIHRTTGNDTGLAVLSMKDKSLVAHLNSTDDISYHLVGNIVNTRSAKLDAWRDFDDVTTVDLASYIRLNHSRLLTSSLVWRPQIFSEVKAQAVSTLKLLYGQINDSLVILKEAPMEAHLALKAIWADARPRIGDFLHDLDDLQVIKDDLDDFETFLNESYSKNDFYVKDIVEFTYYVLDEMAIRNHLESLPGIVNDMWGMMGNTSQSIKQSLTYVVDTIKHAYANFLESVNKALEADFMELVSERLEAMILQYDNFIRDLHMKLLEYWEETWVNATNRLTKYWHELLKSIEPLFFKILHYSESFVFTVWKSLMDFFYNRTQELTDSPYFNYVSTFGHEMDKIYKDLVNNDLITNIKKYSKKLWDAVWTKIEKYIPFKDEIVQLYNEFKDSWQDFLKTPQVVYVREKYQEAYVRLKWWYDYFLIGEALNTIGEIIYVKLTDLAKTALQYEELHRTPKTNFIFDPRVGEIVLEQKLPMSWHAFNSSPDFTEISEYKTVRDFFDEWLTTNKSIWSYYYEIRPYMDLNNLMPPFPGIAMMTEQGALVTYDKQVFTVSERGVFLLTKDYRRDNFTVLMASNDRGTYDLVLVTRTKLVYIDLSKEQVTVSDTVLSLPASVEGLVIDRQMETLSVRGDSGLDVQCNLRFKSCKLGVEGWFYATLGGLLGTYNNEQYDDLQLPNNSFATSVGALATAWRLGPAPPAPAPLPAPPTPHNETQCEKFFQNKVSPFHPCFSVIKAAPFYSECQAGVEACALASAYLELCTQQHVPTHMPDHCNQCTTPHGDVIDEGSFYTLENVPESTDVVFIIEAQVCNKNVRKTKNLDLFLETFDIQLQAGGLTDNRYAVVGFGGRGVYRQPRAVYVNNKVFTDPIEISSHFDTFQIDKLDLARPNKTVKADMFSALNFATKLPFRPGVPRTFLLMPCTRCDSQFMRLDYSSMYQSLMENSVTLHILMDDDFTLSKKRASKYLFGVDSTLAYTNKDYEKLVGDAALRKQVKLPKDKLGLCISLAMETNGTILAGTKLSAERRSARRFSSVAGARAARGAARCARTRCECRAGATHCRPCADTLRMWELAFFSTDDIDDLIDMAMEPPTIPSLS
Summary
Uniprot
S0BCV4
H9JWI0
A0A2A4K630
A0A2W1BAD1
A0A1Z2RRJ0
A0A194Q2X6
+ More
A0A212F5U0 A0A0L7LFN3 A0A2J7QJY6 A0A2J7QJX6 A0A067RG94 A0A139WP32 A0A139WPB9 K7IXR4 A0A182G279 B0WPV2 A0A1Y1LL77 A0A182WA07 A0A2P8Z010 A0A182M2K1 A0A0J7NUE9 A0A151WY88 A0A151I3W2 A0A195D212 A0A026W332 A0A1S4G723 A0A158NZQ3 F4WRK0 Q17BE3 A0A182R2J5 W4VR36 A0A0C9RFN8 A0A1I8M6J1 A0A182QHU6 A0A151IY30 A0A182NT55 A0A182J4N7 A0A182IB32 A0A195F3G0 A0A348G694 A0A1S4GZA4 A0A084VJ90 A0A0M4E168 A0A1I8NQH1 A0A182T296 A0A336LP01 A0A154PLI1 B4KH15 A0A0R3NXZ4 B4G7Z1 Q29NB0 E2C0B0 A0A0M8ZZC0 A0A088AUT9 A0A0Q5WLW8 A0A3B0KBY7 A0A182Y9A7 B3N8G4 W5JC53 B4M9H4 A0A0L0CLX4 A0A2A3E7M2 B3MUX3 A0A1W4W8I7 A0A1W4WA33 Q7KTG2 A0A0J9QYW7 B4JCE5 B4NY63 A0A1A9W006 B4MVA4 A0A1A9V9N0 A0A182F2U2 A0A1B0G0Q6 B4Q7U8 A0A1B0A4Y2 A0A310SMX2 A0A1B0C120 B4HW87 A0A1A9XGW7 A0A1S4H025 E0V9D4 A0A1J1IE84 A0A3L8D672 A0A2H8TTP9 A0A224XHA8 J9K1J6 A0A2S2R693 A0A0A9Y371 A0A0A9XNG6 A0A0A9XUY2 A0A1B6L790 A0A1B6L074 W8B060 A0A1B6G5M2 A0A0A9Y0G8
A0A212F5U0 A0A0L7LFN3 A0A2J7QJY6 A0A2J7QJX6 A0A067RG94 A0A139WP32 A0A139WPB9 K7IXR4 A0A182G279 B0WPV2 A0A1Y1LL77 A0A182WA07 A0A2P8Z010 A0A182M2K1 A0A0J7NUE9 A0A151WY88 A0A151I3W2 A0A195D212 A0A026W332 A0A1S4G723 A0A158NZQ3 F4WRK0 Q17BE3 A0A182R2J5 W4VR36 A0A0C9RFN8 A0A1I8M6J1 A0A182QHU6 A0A151IY30 A0A182NT55 A0A182J4N7 A0A182IB32 A0A195F3G0 A0A348G694 A0A1S4GZA4 A0A084VJ90 A0A0M4E168 A0A1I8NQH1 A0A182T296 A0A336LP01 A0A154PLI1 B4KH15 A0A0R3NXZ4 B4G7Z1 Q29NB0 E2C0B0 A0A0M8ZZC0 A0A088AUT9 A0A0Q5WLW8 A0A3B0KBY7 A0A182Y9A7 B3N8G4 W5JC53 B4M9H4 A0A0L0CLX4 A0A2A3E7M2 B3MUX3 A0A1W4W8I7 A0A1W4WA33 Q7KTG2 A0A0J9QYW7 B4JCE5 B4NY63 A0A1A9W006 B4MVA4 A0A1A9V9N0 A0A182F2U2 A0A1B0G0Q6 B4Q7U8 A0A1B0A4Y2 A0A310SMX2 A0A1B0C120 B4HW87 A0A1A9XGW7 A0A1S4H025 E0V9D4 A0A1J1IE84 A0A3L8D672 A0A2H8TTP9 A0A224XHA8 J9K1J6 A0A2S2R693 A0A0A9Y371 A0A0A9XNG6 A0A0A9XUY2 A0A1B6L790 A0A1B6L074 W8B060 A0A1B6G5M2 A0A0A9Y0G8
Pubmed
23812557
19121390
28756777
28605547
26354079
22118469
+ More
26227816 24845553 18362917 19820115 20075255 26483478 28004739 29403074 24508170 21347285 21719571 17510324 25315136 12364791 24438588 17994087 15632085 20798317 25244985 20920257 23761445 26108605 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17550304 20566863 30249741 25401762 24495485 26823975
26227816 24845553 18362917 19820115 20075255 26483478 28004739 29403074 24508170 21347285 21719571 17510324 25315136 12364791 24438588 17994087 15632085 20798317 25244985 20920257 23761445 26108605 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17550304 20566863 30249741 25401762 24495485 26823975
EMBL
AB700597
BAN58736.1
BABH01018618
BABH01018619
BABH01018620
BABH01018621
+ More
BABH01018622 BABH01018623 BABH01018624 BABH01018625 BABH01018626 NWSH01000142 PCG79120.1 KZ150355 PZC71235.1 KY938823 ASA46464.1 KQ459588 KPI97760.1 AGBW02010140 OWR49093.1 JTDY01001259 KOB74368.1 NEVH01013548 PNF28889.1 PNF28890.1 KK852702 KDR18107.1 KQ971307 KYB29704.1 KYB29707.1 JXUM01039467 KQ561205 KXJ79351.1 DS232030 EDS32540.1 GEZM01055232 JAV73080.1 PYGN01000261 PSN49824.1 AXCM01000090 LBMM01001603 KMQ96030.1 KQ982656 KYQ52727.1 KQ976474 KYM83950.1 KQ976948 KYN06912.1 KK107488 EZA49996.1 ADTU01004889 ADTU01004890 GL888291 EGI63145.1 CH477322 EAT43600.1 GANO01004605 JAB55266.1 GBYB01005798 GBYB01005801 JAG75565.1 JAG75568.1 AXCN02000729 KQ980791 KYN13149.1 APCN01000637 KQ981856 KYN34717.1 FX985636 BBF97967.1 AAAB01008944 ATLV01013476 KE524860 KFB38034.1 CP012523 ALC39054.1 UFQS01000034 UFQT01000034 SSW97913.1 SSX18299.1 KQ434954 KZC12617.1 CH933807 EDW12226.1 CH379060 KRT03824.1 CH479180 EDW28489.1 EAL33432.1 GL451753 EFN78662.1 KQ435794 KOX73997.1 CH954177 KQS70383.1 OUUW01000010 SPP85680.1 EDV58387.2 ADMH02001599 ETN61902.1 CH940654 EDW57850.2 JRES01000209 KNC33266.1 KZ288356 PBC27179.1 CH902624 EDV33038.1 KPU74268.1 AE014134 AAS64667.2 CM002910 KMY89297.1 CH916368 EDW03099.1 CM000157 EDW88665.2 CH963857 EDW76449.2 CCAG010010262 CM000361 EDX04380.1 KQ759866 OAD62482.1 JXJN01023868 CH480818 EDW52282.1 DS234992 EEB09990.1 CVRI01000047 CRK98565.1 QOIP01000012 RLU15967.1 GFXV01005782 MBW17587.1 GFTR01008997 JAW07429.1 ABLF02025947 ABLF02025948 ABLF02025950 ABLF02025951 ABLF02025952 ABLF02025953 ABLF02041568 ABLF02064823 ABLF02065793 GGMS01016261 MBY85464.1 GBHO01017015 JAG26589.1 GBHO01021322 JAG22282.1 GBHO01019870 JAG23734.1 GEBQ01020388 JAT19589.1 GEBQ01022875 JAT17102.1 GAMC01016087 JAB90468.1 GECZ01012035 JAS57734.1 GBHO01017037 GDHC01021459 JAG26567.1 JAP97169.1
BABH01018622 BABH01018623 BABH01018624 BABH01018625 BABH01018626 NWSH01000142 PCG79120.1 KZ150355 PZC71235.1 KY938823 ASA46464.1 KQ459588 KPI97760.1 AGBW02010140 OWR49093.1 JTDY01001259 KOB74368.1 NEVH01013548 PNF28889.1 PNF28890.1 KK852702 KDR18107.1 KQ971307 KYB29704.1 KYB29707.1 JXUM01039467 KQ561205 KXJ79351.1 DS232030 EDS32540.1 GEZM01055232 JAV73080.1 PYGN01000261 PSN49824.1 AXCM01000090 LBMM01001603 KMQ96030.1 KQ982656 KYQ52727.1 KQ976474 KYM83950.1 KQ976948 KYN06912.1 KK107488 EZA49996.1 ADTU01004889 ADTU01004890 GL888291 EGI63145.1 CH477322 EAT43600.1 GANO01004605 JAB55266.1 GBYB01005798 GBYB01005801 JAG75565.1 JAG75568.1 AXCN02000729 KQ980791 KYN13149.1 APCN01000637 KQ981856 KYN34717.1 FX985636 BBF97967.1 AAAB01008944 ATLV01013476 KE524860 KFB38034.1 CP012523 ALC39054.1 UFQS01000034 UFQT01000034 SSW97913.1 SSX18299.1 KQ434954 KZC12617.1 CH933807 EDW12226.1 CH379060 KRT03824.1 CH479180 EDW28489.1 EAL33432.1 GL451753 EFN78662.1 KQ435794 KOX73997.1 CH954177 KQS70383.1 OUUW01000010 SPP85680.1 EDV58387.2 ADMH02001599 ETN61902.1 CH940654 EDW57850.2 JRES01000209 KNC33266.1 KZ288356 PBC27179.1 CH902624 EDV33038.1 KPU74268.1 AE014134 AAS64667.2 CM002910 KMY89297.1 CH916368 EDW03099.1 CM000157 EDW88665.2 CH963857 EDW76449.2 CCAG010010262 CM000361 EDX04380.1 KQ759866 OAD62482.1 JXJN01023868 CH480818 EDW52282.1 DS234992 EEB09990.1 CVRI01000047 CRK98565.1 QOIP01000012 RLU15967.1 GFXV01005782 MBW17587.1 GFTR01008997 JAW07429.1 ABLF02025947 ABLF02025948 ABLF02025950 ABLF02025951 ABLF02025952 ABLF02025953 ABLF02041568 ABLF02064823 ABLF02065793 GGMS01016261 MBY85464.1 GBHO01017015 JAG26589.1 GBHO01021322 JAG22282.1 GBHO01019870 JAG23734.1 GEBQ01020388 JAT19589.1 GEBQ01022875 JAT17102.1 GAMC01016087 JAB90468.1 GECZ01012035 JAS57734.1 GBHO01017037 GDHC01021459 JAG26567.1 JAP97169.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000037510
UP000235965
+ More
UP000027135 UP000007266 UP000002358 UP000069940 UP000249989 UP000002320 UP000075920 UP000245037 UP000075883 UP000036403 UP000075809 UP000078540 UP000078542 UP000053097 UP000005205 UP000007755 UP000008820 UP000075900 UP000095301 UP000075886 UP000078492 UP000075884 UP000075880 UP000075840 UP000078541 UP000030765 UP000092553 UP000095300 UP000075901 UP000076502 UP000009192 UP000001819 UP000008744 UP000008237 UP000053105 UP000005203 UP000008711 UP000268350 UP000076408 UP000000673 UP000008792 UP000037069 UP000242457 UP000007801 UP000192221 UP000000803 UP000001070 UP000002282 UP000091820 UP000007798 UP000078200 UP000069272 UP000092444 UP000000304 UP000092445 UP000092460 UP000001292 UP000092443 UP000009046 UP000183832 UP000279307 UP000007819
UP000027135 UP000007266 UP000002358 UP000069940 UP000249989 UP000002320 UP000075920 UP000245037 UP000075883 UP000036403 UP000075809 UP000078540 UP000078542 UP000053097 UP000005205 UP000007755 UP000008820 UP000075900 UP000095301 UP000075886 UP000078492 UP000075884 UP000075880 UP000075840 UP000078541 UP000030765 UP000092553 UP000095300 UP000075901 UP000076502 UP000009192 UP000001819 UP000008744 UP000008237 UP000053105 UP000005203 UP000008711 UP000268350 UP000076408 UP000000673 UP000008792 UP000037069 UP000242457 UP000007801 UP000192221 UP000000803 UP000001070 UP000002282 UP000091820 UP000007798 UP000078200 UP000069272 UP000092444 UP000000304 UP000092445 UP000092460 UP000001292 UP000092443 UP000009046 UP000183832 UP000279307 UP000007819
Pfam
Interpro
IPR001747
Lipid_transpt_N
+ More
IPR011030 Lipovitellin_superhlx_dom
IPR001846 VWF_type-D
IPR015819 Lipid_transp_b-sht_shell
IPR015816 Vitellinogen_b-sht_N
IPR015255 Vitellinogen_open_b-sht
IPR009454 Lipid_transpt_open_b-sht
IPR002035 VWF_A
IPR036465 vWFA_dom_sf
IPR015817 Vitellinogen_open_b-sht_sub1
IPR014853 Unchr_dom_Cys-rich
IPR005027 Glyco_trans_43
IPR029044 Nucleotide-diphossugar_trans
IPR011030 Lipovitellin_superhlx_dom
IPR001846 VWF_type-D
IPR015819 Lipid_transp_b-sht_shell
IPR015816 Vitellinogen_b-sht_N
IPR015255 Vitellinogen_open_b-sht
IPR009454 Lipid_transpt_open_b-sht
IPR002035 VWF_A
IPR036465 vWFA_dom_sf
IPR015817 Vitellinogen_open_b-sht_sub1
IPR014853 Unchr_dom_Cys-rich
IPR005027 Glyco_trans_43
IPR029044 Nucleotide-diphossugar_trans
CDD
ProteinModelPortal
S0BCV4
H9JWI0
A0A2A4K630
A0A2W1BAD1
A0A1Z2RRJ0
A0A194Q2X6
+ More
A0A212F5U0 A0A0L7LFN3 A0A2J7QJY6 A0A2J7QJX6 A0A067RG94 A0A139WP32 A0A139WPB9 K7IXR4 A0A182G279 B0WPV2 A0A1Y1LL77 A0A182WA07 A0A2P8Z010 A0A182M2K1 A0A0J7NUE9 A0A151WY88 A0A151I3W2 A0A195D212 A0A026W332 A0A1S4G723 A0A158NZQ3 F4WRK0 Q17BE3 A0A182R2J5 W4VR36 A0A0C9RFN8 A0A1I8M6J1 A0A182QHU6 A0A151IY30 A0A182NT55 A0A182J4N7 A0A182IB32 A0A195F3G0 A0A348G694 A0A1S4GZA4 A0A084VJ90 A0A0M4E168 A0A1I8NQH1 A0A182T296 A0A336LP01 A0A154PLI1 B4KH15 A0A0R3NXZ4 B4G7Z1 Q29NB0 E2C0B0 A0A0M8ZZC0 A0A088AUT9 A0A0Q5WLW8 A0A3B0KBY7 A0A182Y9A7 B3N8G4 W5JC53 B4M9H4 A0A0L0CLX4 A0A2A3E7M2 B3MUX3 A0A1W4W8I7 A0A1W4WA33 Q7KTG2 A0A0J9QYW7 B4JCE5 B4NY63 A0A1A9W006 B4MVA4 A0A1A9V9N0 A0A182F2U2 A0A1B0G0Q6 B4Q7U8 A0A1B0A4Y2 A0A310SMX2 A0A1B0C120 B4HW87 A0A1A9XGW7 A0A1S4H025 E0V9D4 A0A1J1IE84 A0A3L8D672 A0A2H8TTP9 A0A224XHA8 J9K1J6 A0A2S2R693 A0A0A9Y371 A0A0A9XNG6 A0A0A9XUY2 A0A1B6L790 A0A1B6L074 W8B060 A0A1B6G5M2 A0A0A9Y0G8
A0A212F5U0 A0A0L7LFN3 A0A2J7QJY6 A0A2J7QJX6 A0A067RG94 A0A139WP32 A0A139WPB9 K7IXR4 A0A182G279 B0WPV2 A0A1Y1LL77 A0A182WA07 A0A2P8Z010 A0A182M2K1 A0A0J7NUE9 A0A151WY88 A0A151I3W2 A0A195D212 A0A026W332 A0A1S4G723 A0A158NZQ3 F4WRK0 Q17BE3 A0A182R2J5 W4VR36 A0A0C9RFN8 A0A1I8M6J1 A0A182QHU6 A0A151IY30 A0A182NT55 A0A182J4N7 A0A182IB32 A0A195F3G0 A0A348G694 A0A1S4GZA4 A0A084VJ90 A0A0M4E168 A0A1I8NQH1 A0A182T296 A0A336LP01 A0A154PLI1 B4KH15 A0A0R3NXZ4 B4G7Z1 Q29NB0 E2C0B0 A0A0M8ZZC0 A0A088AUT9 A0A0Q5WLW8 A0A3B0KBY7 A0A182Y9A7 B3N8G4 W5JC53 B4M9H4 A0A0L0CLX4 A0A2A3E7M2 B3MUX3 A0A1W4W8I7 A0A1W4WA33 Q7KTG2 A0A0J9QYW7 B4JCE5 B4NY63 A0A1A9W006 B4MVA4 A0A1A9V9N0 A0A182F2U2 A0A1B0G0Q6 B4Q7U8 A0A1B0A4Y2 A0A310SMX2 A0A1B0C120 B4HW87 A0A1A9XGW7 A0A1S4H025 E0V9D4 A0A1J1IE84 A0A3L8D672 A0A2H8TTP9 A0A224XHA8 J9K1J6 A0A2S2R693 A0A0A9Y371 A0A0A9XNG6 A0A0A9XUY2 A0A1B6L790 A0A1B6L074 W8B060 A0A1B6G5M2 A0A0A9Y0G8
Ontologies
GO
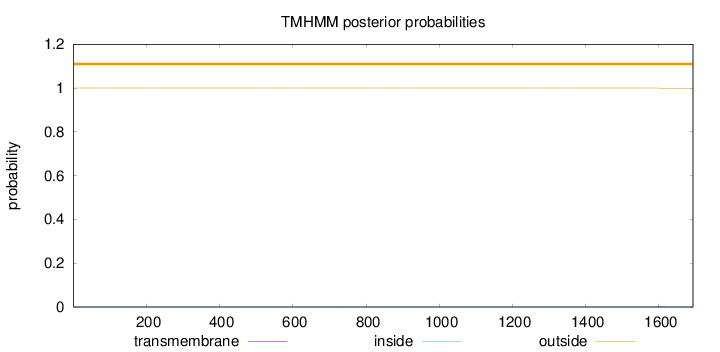
Topology
Length:
1693
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01166
Exp number, first 60 AAs:
0.00122
Total prob of N-in:
0.00010
outside
1 - 1693
Population Genetic Test Statistics
Pi
253.924479
Theta
157.739812
Tajima's D
1.802145
CLR
0.396373
CSRT
0.846507674616269
Interpretation
Uncertain
