Gene
KWMTBOMO16267 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013895
Annotation
5'-methylthioadenosine_phosphorylase_[Bombyx_mori]
Full name
S-methyl-5'-thioadenosine phosphorylase
+ More
Purine nucleoside phosphorylase
Purine nucleoside phosphorylase
Alternative Name
5'-methylthioadenosine phosphorylase
Location in the cell
Cytoplasmic Reliability : 2.21
Sequence
CDS
ATGGGCGATGTAAGAAAAATAAAGATCGGAATTATCGGAGGTTCGGGCTTCGATGATCCGACGCTTTTCGAGAATCAAATCGAAAAGGAGGTGGTCACACCGTTTGGGAGACCCTCGGACGTACTGATAGAGGGTCAAATCAAACGGGTCCAATGCGTTCTGCTGGCACGACACGGCAGAAAACATCAACTTCAACCTAGCGATGTGAACTATCGAGCTAATATTTGGGCACTGAAGCAAGTCGGCTGCACTCACATCCTCGCCACGACCGCCACTGGGTCTCTAGTTGAAGAATACAGGCCTGGGGATCTGGTCATATTGGACGATTTCATTGATAGGACTTGGGGTCGGAAGTGCACGTTTTACGACAACACGGAGGGGGGTCCGCGCGGCGTGTGCCACCTGCCCATGCGGCCGGCGTATTGTGGGAGAGCGCGCGCCGCACTGTACAGCGCGGCCAAGTCTAGGGGGTACTCGTGCCATGAAACCGGCACTGCGGTTGTCATACAGGGACCCAGATTCTCAAGTCGCGCCGAAAGCTTGGTCCATAGACAATGGGGTGGCCATCTTGTGAACATGACAACTGTCCCTGAGGTTGTGTTAGCTAAAGAAGCCGGCTTGAGTTACGCTGCCGTGGCACTAGTCACAGATTACGATTGTTGGAGGGAAAATGAAAAATCCGTCTCGGTGAGCGAAGTTCTGGCAACATTCTCGAAGAACGTTAAGAAGGCTGCGGATGTTATCGTAGACGCAGTTCAGATATTAGGAGCCGATACCGACCTCCAATATCTGGACGATCACCAGGAACTCGTCAAATCCGCCATAATGCTCAAGGACTGA
Protein
MGDVRKIKIGIIGGSGFDDPTLFENQIEKEVVTPFGRPSDVLIEGQIKRVQCVLLARHGRKHQLQPSDVNYRANIWALKQVGCTHILATTATGSLVEEYRPGDLVILDDFIDRTWGRKCTFYDNTEGGPRGVCHLPMRPAYCGRARAALYSAAKSRGYSCHETGTAVVIQGPRFSSRAESLVHRQWGGHLVNMTTVPEVVLAKEAGLSYAAVALVTDYDCWRENEKSVSVSEVLATFSKNVKKAADVIVDAVQILGADTDLQYLDDHQELVKSAIMLKD
Summary
Description
Catalyzes the reversible phosphorylation of S-methyl-5'-thioadenosine (MTA) to adenine and 5-methylthioribose-1-phosphate. Involved in the breakdown of MTA, a major by-product of polyamine biosynthesis. Responsible for the first step in the methionine salvage pathway after MTA has been generated from S-adenosylmethionine. Has broad substrate specificity with 6-aminopurine nucleosides as preferred substrates.
Purine nucleoside phosphorylase involved in purine salvage.
Purine nucleoside phosphorylase involved in purine salvage.
Catalytic Activity
phosphate + S-methyl-5'-thioadenosine = adenine + S-methyl-5-thio-alpha-D-ribose 1-phosphate
a purine D-ribonucleoside + phosphate = a purine nucleobase + alpha-D-ribose 1-phosphate
a purine D-ribonucleoside + phosphate = a purine nucleobase + alpha-D-ribose 1-phosphate
Subunit
Homotrimer.
Miscellaneous
Although this enzyme belongs to the family of MTA phosphorylases based on sequence homology, it lacks several conserved amino acids in the substrate binding pocket that confer specificity towards MTA.
Similarity
Belongs to the PNP/MTAP phosphorylase family. MTAP subfamily.
Keywords
Complete proteome
Cytoplasm
Glycosyltransferase
Nucleus
Purine salvage
Reference proteome
Transferase
Acetylation
Feature
chain S-methyl-5'-thioadenosine phosphorylase
Uniprot
Q1HQ82
H9JWI1
A0A2A4K4C7
A0A2W1B6H2
I4DLI4
A0A0N1PH14
+ More
A0A194PWJ7 A0A212FEM5 A0A220K8J9 S4PSF7 A0A3S2M6L3 A0A336LVV3 D6WPA2 A0A1L8DXQ8 T1P8V4 A0A1I8N9R6 V5GMI9 A0A1Y1MEH6 A0A1W4XN90 E3TFK1 A0A093Q5T8 B2GQM6 Q7ZV22 A0A3B3ZNI1 F1NCV7 A0A2I4D6S4 A0A3Q0T2T2 A0A091IS19 A0A3B3BKU3 A0A3P9LHK0 A0A093IKE2 A0A087RD82 A0A182M5A4 H0WX98 A0A182SMW3 G1T2Z8 A0A182N801 A0A182PHE2 A0A182YNS0 F7AA57 A0A093I3N3 A0A3B4Z2M5 A0A2U3ZTG6 A0A091V1J1 A0A087VKB9 A0A1W4YT55 A0A2U3WU82 G3T3Y4 A0A099ZW69 A0A091VGE7 A0A091L1G8 A0A182VIQ8 A0A182K140 A0A182LBD4 A0A182XJA1 A0A182HP61 Q7Q9N9 A0A3L7I8P3 Q9CQ65 U6CT37 A0A2Y9L3N6 M3YX12 A0A182RTP8 A0A091MTI6 A0A091P836 A0A1B6CHF1 A0A093DEC8 A0A093FXA3 A0A091ERY4 A0A3B4XVF1 A0A093P8F8 I3KLX3 A0A2U3YP82 A0A383ZPP0 A0A3B4TAR6 A0A3Q0CS45 Q6FHT1 A0A3B4FPQ9 A0A3P8QNY6 A0A1A8PQS3 A0A1A8MHI1 G1LLW4 A0A3S2PTX5 A0A1A8F492 A0A2K6DWP1 A0A2I2UPB1 F6RQM9 I7GJ70 A0A3P9CWM5 A0A3Q7MNR7 A0A1I7Q457 A0A250YEG0 A0A099ZIL1 A0A3Q1ING3 Q5ZHQ7 A0A093DXQ9 A0A0G2K583 A0A0A1X9J3 A0A2K6M620
A0A194PWJ7 A0A212FEM5 A0A220K8J9 S4PSF7 A0A3S2M6L3 A0A336LVV3 D6WPA2 A0A1L8DXQ8 T1P8V4 A0A1I8N9R6 V5GMI9 A0A1Y1MEH6 A0A1W4XN90 E3TFK1 A0A093Q5T8 B2GQM6 Q7ZV22 A0A3B3ZNI1 F1NCV7 A0A2I4D6S4 A0A3Q0T2T2 A0A091IS19 A0A3B3BKU3 A0A3P9LHK0 A0A093IKE2 A0A087RD82 A0A182M5A4 H0WX98 A0A182SMW3 G1T2Z8 A0A182N801 A0A182PHE2 A0A182YNS0 F7AA57 A0A093I3N3 A0A3B4Z2M5 A0A2U3ZTG6 A0A091V1J1 A0A087VKB9 A0A1W4YT55 A0A2U3WU82 G3T3Y4 A0A099ZW69 A0A091VGE7 A0A091L1G8 A0A182VIQ8 A0A182K140 A0A182LBD4 A0A182XJA1 A0A182HP61 Q7Q9N9 A0A3L7I8P3 Q9CQ65 U6CT37 A0A2Y9L3N6 M3YX12 A0A182RTP8 A0A091MTI6 A0A091P836 A0A1B6CHF1 A0A093DEC8 A0A093FXA3 A0A091ERY4 A0A3B4XVF1 A0A093P8F8 I3KLX3 A0A2U3YP82 A0A383ZPP0 A0A3B4TAR6 A0A3Q0CS45 Q6FHT1 A0A3B4FPQ9 A0A3P8QNY6 A0A1A8PQS3 A0A1A8MHI1 G1LLW4 A0A3S2PTX5 A0A1A8F492 A0A2K6DWP1 A0A2I2UPB1 F6RQM9 I7GJ70 A0A3P9CWM5 A0A3Q7MNR7 A0A1I7Q457 A0A250YEG0 A0A099ZIL1 A0A3Q1ING3 Q5ZHQ7 A0A093DXQ9 A0A0G2K583 A0A0A1X9J3 A0A2K6M620
EC Number
2.4.2.28
2.4.2.1
2.4.2.1
Pubmed
19121390
28756777
22651552
26354079
22118469
28630352
+ More
23622113 18362917 19820115 25315136 28004739 20634964 23594743 25463417 15592404 29451363 17554307 21993624 25244985 17495919 20966253 12364791 29704459 12824877 16141072 15489334 21183079 23806337 25186727 26319212 28071753 20010809 17975172 17431167 17194215 28087693 15642098 15057822 25830018
23622113 18362917 19820115 25315136 28004739 20634964 23594743 25463417 15592404 29451363 17554307 21993624 25244985 17495919 20966253 12364791 29704459 12824877 16141072 15489334 21183079 23806337 25186727 26319212 28071753 20010809 17975172 17431167 17194215 28087693 15642098 15057822 25830018
EMBL
DQ443170
ABF51259.1
BABH01018616
BABH01018617
NWSH01000142
PCG79115.1
+ More
KZ150355 PZC71238.1 AK402152 BAM18774.1 KQ460417 KPJ14874.1 KQ459588 KPI97746.1 AGBW02008913 OWR52191.1 MF319699 ASJ26413.1 GAIX01013868 JAA78692.1 RSAL01000025 RVE52156.1 UFQS01000015 UFQT01000015 SSW97362.1 SSX17748.1 KQ971354 EFA07386.2 GFDF01002890 JAV11194.1 KA645206 AFP59835.1 GALX01005644 JAB62822.1 GEZM01036390 JAV82735.1 GU589135 ADO29087.1 KL416278 KFW81805.1 BC164800 AAI64800.1 BX323448 BC046035 BC056545 AC195266 KL218126 KFP02436.1 KK589556 KFV99243.1 KL226301 KFM11436.1 AXCM01001928 AAQR03014015 AAQR03014016 AAQR03014017 AAQR03014018 AAQR03014019 AAQR03014020 AAGW02033076 AAGW02033077 AAGW02033078 AAGW02033079 KL214741 KFV61351.1 KK409286 KFQ83628.1 KL496516 KFO13061.1 KL869929 KGL86529.1 KK733937 KFR01538.1 KL291445 KFP49637.1 APCN01002921 AAAB01008900 RAZU01000092 RLQ73941.1 AB056100 AK005064 AK011421 AK167319 BC003858 HAAF01001820 CCP73646.1 AEYP01053750 KK837640 KFP80823.1 KL381615 KFP87675.1 GEDC01024439 JAS12859.1 KN125705 KFU83838.1 KK399278 KFV59011.1 KK718913 KFO59781.1 KL225291 KFW70535.1 AERX01027956 AERX01027957 FJ224338 CR541670 ACI46030.1 CAG46471.1 HAEH01008106 HAEI01009746 SBR83603.1 HAEF01014660 HAEG01003409 SBR55819.1 ACTA01169101 ACTA01177099 CM012437 RVE76207.1 HAEB01007633 HAEC01010783 SBQ54160.1 AANG04001372 JSUE03014969 JSUE03014970 AQIA01024519 AQIA01024520 AQIA01024521 AQIA01024522 AB174101 BAE91163.1 GFFW01002775 JAV42013.1 KL893441 KGL80818.1 AJ721077 CAG32736.1 KL452767 KFV06966.1 AABR07049152 GBXI01006897 JAD07395.1
KZ150355 PZC71238.1 AK402152 BAM18774.1 KQ460417 KPJ14874.1 KQ459588 KPI97746.1 AGBW02008913 OWR52191.1 MF319699 ASJ26413.1 GAIX01013868 JAA78692.1 RSAL01000025 RVE52156.1 UFQS01000015 UFQT01000015 SSW97362.1 SSX17748.1 KQ971354 EFA07386.2 GFDF01002890 JAV11194.1 KA645206 AFP59835.1 GALX01005644 JAB62822.1 GEZM01036390 JAV82735.1 GU589135 ADO29087.1 KL416278 KFW81805.1 BC164800 AAI64800.1 BX323448 BC046035 BC056545 AC195266 KL218126 KFP02436.1 KK589556 KFV99243.1 KL226301 KFM11436.1 AXCM01001928 AAQR03014015 AAQR03014016 AAQR03014017 AAQR03014018 AAQR03014019 AAQR03014020 AAGW02033076 AAGW02033077 AAGW02033078 AAGW02033079 KL214741 KFV61351.1 KK409286 KFQ83628.1 KL496516 KFO13061.1 KL869929 KGL86529.1 KK733937 KFR01538.1 KL291445 KFP49637.1 APCN01002921 AAAB01008900 RAZU01000092 RLQ73941.1 AB056100 AK005064 AK011421 AK167319 BC003858 HAAF01001820 CCP73646.1 AEYP01053750 KK837640 KFP80823.1 KL381615 KFP87675.1 GEDC01024439 JAS12859.1 KN125705 KFU83838.1 KK399278 KFV59011.1 KK718913 KFO59781.1 KL225291 KFW70535.1 AERX01027956 AERX01027957 FJ224338 CR541670 ACI46030.1 CAG46471.1 HAEH01008106 HAEI01009746 SBR83603.1 HAEF01014660 HAEG01003409 SBR55819.1 ACTA01169101 ACTA01177099 CM012437 RVE76207.1 HAEB01007633 HAEC01010783 SBQ54160.1 AANG04001372 JSUE03014969 JSUE03014970 AQIA01024519 AQIA01024520 AQIA01024521 AQIA01024522 AB174101 BAE91163.1 GFFW01002775 JAV42013.1 KL893441 KGL80818.1 AJ721077 CAG32736.1 KL452767 KFV06966.1 AABR07049152 GBXI01006897 JAD07395.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000283053
+ More
UP000007266 UP000095301 UP000192223 UP000221080 UP000000437 UP000261520 UP000000539 UP000192220 UP000261340 UP000054308 UP000261560 UP000265180 UP000265200 UP000053286 UP000075883 UP000005225 UP000075901 UP000001811 UP000075884 UP000075885 UP000076408 UP000002280 UP000053875 UP000261400 UP000245340 UP000192224 UP000007646 UP000053858 UP000053605 UP000075903 UP000075881 UP000075882 UP000076407 UP000075840 UP000007062 UP000273346 UP000000589 UP000248482 UP000000715 UP000075900 UP000052976 UP000261360 UP000054081 UP000005207 UP000245341 UP000261681 UP000261420 UP000189706 UP000261460 UP000265100 UP000008912 UP000233120 UP000011712 UP000006718 UP000233100 UP000265160 UP000286641 UP000053641 UP000265040 UP000002494 UP000233180
UP000007266 UP000095301 UP000192223 UP000221080 UP000000437 UP000261520 UP000000539 UP000192220 UP000261340 UP000054308 UP000261560 UP000265180 UP000265200 UP000053286 UP000075883 UP000005225 UP000075901 UP000001811 UP000075884 UP000075885 UP000076408 UP000002280 UP000053875 UP000261400 UP000245340 UP000192224 UP000007646 UP000053858 UP000053605 UP000075903 UP000075881 UP000075882 UP000076407 UP000075840 UP000007062 UP000273346 UP000000589 UP000248482 UP000000715 UP000075900 UP000052976 UP000261360 UP000054081 UP000005207 UP000245341 UP000261681 UP000261420 UP000189706 UP000261460 UP000265100 UP000008912 UP000233120 UP000011712 UP000006718 UP000233100 UP000265160 UP000286641 UP000053641 UP000265040 UP000002494 UP000233180
Interpro
SUPFAM
SSF53167
SSF53167
Gene 3D
ProteinModelPortal
Q1HQ82
H9JWI1
A0A2A4K4C7
A0A2W1B6H2
I4DLI4
A0A0N1PH14
+ More
A0A194PWJ7 A0A212FEM5 A0A220K8J9 S4PSF7 A0A3S2M6L3 A0A336LVV3 D6WPA2 A0A1L8DXQ8 T1P8V4 A0A1I8N9R6 V5GMI9 A0A1Y1MEH6 A0A1W4XN90 E3TFK1 A0A093Q5T8 B2GQM6 Q7ZV22 A0A3B3ZNI1 F1NCV7 A0A2I4D6S4 A0A3Q0T2T2 A0A091IS19 A0A3B3BKU3 A0A3P9LHK0 A0A093IKE2 A0A087RD82 A0A182M5A4 H0WX98 A0A182SMW3 G1T2Z8 A0A182N801 A0A182PHE2 A0A182YNS0 F7AA57 A0A093I3N3 A0A3B4Z2M5 A0A2U3ZTG6 A0A091V1J1 A0A087VKB9 A0A1W4YT55 A0A2U3WU82 G3T3Y4 A0A099ZW69 A0A091VGE7 A0A091L1G8 A0A182VIQ8 A0A182K140 A0A182LBD4 A0A182XJA1 A0A182HP61 Q7Q9N9 A0A3L7I8P3 Q9CQ65 U6CT37 A0A2Y9L3N6 M3YX12 A0A182RTP8 A0A091MTI6 A0A091P836 A0A1B6CHF1 A0A093DEC8 A0A093FXA3 A0A091ERY4 A0A3B4XVF1 A0A093P8F8 I3KLX3 A0A2U3YP82 A0A383ZPP0 A0A3B4TAR6 A0A3Q0CS45 Q6FHT1 A0A3B4FPQ9 A0A3P8QNY6 A0A1A8PQS3 A0A1A8MHI1 G1LLW4 A0A3S2PTX5 A0A1A8F492 A0A2K6DWP1 A0A2I2UPB1 F6RQM9 I7GJ70 A0A3P9CWM5 A0A3Q7MNR7 A0A1I7Q457 A0A250YEG0 A0A099ZIL1 A0A3Q1ING3 Q5ZHQ7 A0A093DXQ9 A0A0G2K583 A0A0A1X9J3 A0A2K6M620
A0A194PWJ7 A0A212FEM5 A0A220K8J9 S4PSF7 A0A3S2M6L3 A0A336LVV3 D6WPA2 A0A1L8DXQ8 T1P8V4 A0A1I8N9R6 V5GMI9 A0A1Y1MEH6 A0A1W4XN90 E3TFK1 A0A093Q5T8 B2GQM6 Q7ZV22 A0A3B3ZNI1 F1NCV7 A0A2I4D6S4 A0A3Q0T2T2 A0A091IS19 A0A3B3BKU3 A0A3P9LHK0 A0A093IKE2 A0A087RD82 A0A182M5A4 H0WX98 A0A182SMW3 G1T2Z8 A0A182N801 A0A182PHE2 A0A182YNS0 F7AA57 A0A093I3N3 A0A3B4Z2M5 A0A2U3ZTG6 A0A091V1J1 A0A087VKB9 A0A1W4YT55 A0A2U3WU82 G3T3Y4 A0A099ZW69 A0A091VGE7 A0A091L1G8 A0A182VIQ8 A0A182K140 A0A182LBD4 A0A182XJA1 A0A182HP61 Q7Q9N9 A0A3L7I8P3 Q9CQ65 U6CT37 A0A2Y9L3N6 M3YX12 A0A182RTP8 A0A091MTI6 A0A091P836 A0A1B6CHF1 A0A093DEC8 A0A093FXA3 A0A091ERY4 A0A3B4XVF1 A0A093P8F8 I3KLX3 A0A2U3YP82 A0A383ZPP0 A0A3B4TAR6 A0A3Q0CS45 Q6FHT1 A0A3B4FPQ9 A0A3P8QNY6 A0A1A8PQS3 A0A1A8MHI1 G1LLW4 A0A3S2PTX5 A0A1A8F492 A0A2K6DWP1 A0A2I2UPB1 F6RQM9 I7GJ70 A0A3P9CWM5 A0A3Q7MNR7 A0A1I7Q457 A0A250YEG0 A0A099ZIL1 A0A3Q1ING3 Q5ZHQ7 A0A093DXQ9 A0A0G2K583 A0A0A1X9J3 A0A2K6M620
PDB
6DZ3
E-value=5.06313e-81,
Score=765
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
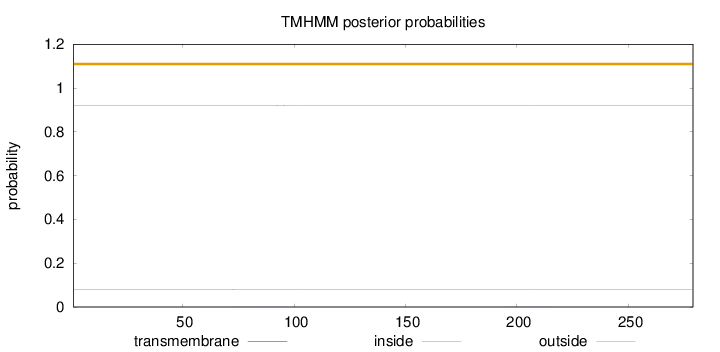
Length:
279
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0195
Exp number, first 60 AAs:
0.00014
Total prob of N-in:
0.07922
outside
1 - 279
Population Genetic Test Statistics
Pi
173.888615
Theta
165.857413
Tajima's D
0.269888
CLR
175.908642
CSRT
0.448177591120444
Interpretation
Uncertain
