Gene
KWMTBOMO16265
Pre Gene Modal
BGIBMGA013997
Annotation
PREDICTED:_uncharacterized_protein_LOC105842614_[Bombyx_mori]
Full name
ATP-dependent DNA helicase
Location in the cell
Nuclear Reliability : 3.816
Sequence
CDS
ATGGCCTTCGAGCAGCAGAGCGCCGCCGCCATGCAGGCTCTCCAGCATCAGCTGCTCAGAGGTCTGAGCGGCGCGGGCGGGTCGGGCGGCATGTCGGCGGCCGCGGCCATGATGCAGTTCACTCCGCTGCTGTACTCCTACCAGCTCGCCATGGCGCAGGCCTCCGCGCTCGGCAAGAGATATTCAATGATGGAAGAATCCACAAATCCTGAGGAATCTTCGAATGCAATTACTATACAAGTTGATGTCCATCATCGGCATTATTCACAACTCAGTGCATCAATCGCTACCACAAGCCATGACTGTGTGAATGAACCTGAAACAACGAGTGATCTTCCTTCAATCCACCGTGATTATCCAAAAACAGCTGCACAGCGAATGGCAGAGTATAGAGCTCGGAAAAAAAAGGAAACTCCAATAGTGAATCATCGAAAACATAAAAAATCAGACGCAGAGCGTGCAAAAGAGTATAGAGCCCGAAAAAAAGCGAAGATCCAATCAGCAGACGCAGAGCGTGCAAAAGAGTATAGAGCCCGAAACAAGGCTCAAATCCAAGAACACACAAGCCAAAGTCAGAACTTTGTTGAGTCAACATCAATTCAACAATCAGAACATGAAACTAATCATCTGGCTTACACGGATTTTCATCGTCATCATCTGGCTCATGATGAATTTCAAAAGAAATTTGTTCGAAATTCATTTGGTTATGTGTGCTCAGTTTGTGAAAGACTGTGGTTTAAAGATGACTTACGATCTGCATCCCTTCAACATAATGCAATCTTGAAAACAATAGTTCCTCATCTTGATATAAAGGACGTTGCTCTCTGTAATACCTGTGTAGCATCTTTAAATAATAATAGTATTCCTGTGATGGCTTCTTATAATGGATTCAAATTTCCGAAGAAGCCCGATTATTTACCGCCATTAGACTTAGTATCTGAACGATTGATTTCTCCGCGAATCCCATTCATGCAGATTAGGAGATTAAGACACGTTAACGGTCAGTATGGTATTTATGGACAAATAATCAATGTTCCAGTGTCTATTGATTCAATGGTTCGTACTCTCCCCAGAAGTTTATCTGATGATCACTGTATCAATGTGCACATTAAACGCAAGAAAATCCATAAATCGAGTTATTTGCATGGTATAATAAACAAAGGTACAATAAAGATTTGGCTTCAATTTTTATTAAAGTCGCCATTATATACTATGTATGATATCAAAATTGATGAATCCTTTTTCGAAAACAATGACATTGATACTATACCTCAAGATGAAATAAGCGAACACGTACCAATCGAAGAAAGTCTGGTATCTCAGCAGCAAACATTGATGTGGAATGAAGAAAAATATTTAAGATTAGCCCCAGGAGAAAAAAATCTTCCACATAGTTTACTTTTTGATGAACACGCGGAGGAATTGTCTTTTCCTGCAATTTATTTGGGCCAATTTCGTACGTTTCGTGATGGAGTACGAGTAACTCCATATACAATGGCTACAAGTGAACTTAGACGCTCGGACCGTCGTGCTGTTACTCCACATCATGTATTATACGTGGCAATGAAAATAATGAGACTTCGAGTTCGTGATTCATTAACAGTTGCCTTCAAACATATAGGAAAAGACACAAAAATCACACGTGAACAAATTGAAAATGAAGATTATGTCAACAATTGCATTGAAACTAATTTTGCATTTCTGAAATCAATTCCCAATTCATTATGGTACTGGGCTGATAAAAAGAAAAATTTATTTGCAATGATTAGACAGCTAGGTAAACCAACAATATTTCTCACTATTAGTGCAAATGAAATTGGATGGACTGATTTACTTCAATTATTATATAAGTTGAAAAATAATGGAGCCGAATTATCAAAAAAAGATGCTGAAGAATTACACTATCTTGAAAAAGTTACTCTGATTAATGAAGATTCTGTAACGTGCGCAATCTATTTTAATAAACTGGTAAACGTACTCTTGAATATATTACAGTCTCCTAAGAAGAGTCCGTTTGGAAAATATTATGTGATGCATTATTTCAAAAGAATTGAATTTCAGCACCGAGGTAGTCCACATGCCCATACATTACTGTGGCTTGCAAATGCACCGAATGATGCTTTAGGCGAGAATAAAATGGATGCAATTTCACTGATTAATCATCTTATCTCGATTTCATCGAAAGAAGCATCAGAGCATATAAAATTACAAACACACAAGCACTCATTCACCTGTTACAAAAGAATATCGGCTCATAGACCTCAACAATGTCGTTTTGAAGCTCCATTCTTGCCTTCTCGTTCTACAGAAATTTTATTACCTATCCAACAGGATGAAATTGGTTTTAATCGTTATTTGCAATCTTACAGAAATATTCGAATTAATCTCGAAAATAATGATTATGAGGATTTTGATAGTTTTTATCAGCAAAATAATATTTCATCAGATGATGAATATCGCAATATACTTCGAGCGGGAATCAAGAGACCAAGAGTTTTTATAAAACGCGAACCCAGTGAGAAATGGCATAATCCATTTAATCCGTTTATTCTGAATCTGGTAAAATCGAATATGGACATCCAGTTCATAACAGAGGAATATTCGTGTGCCCAGTATGTAGCTGAGTATGTCAATAAAACAAATCGCGGAATAAGTAATTTACAACGTGAAATCATCAAATGTATAGATGAACACCCGGAGTTCGATGTTGTGGAAATCACAAGAAAATTGGGAGTTGAGATGCTGAACAGTGTTGAAATACCAAATCAAGAAGCTGCCTGGTATCTTTTAAGAGAGCCGATGTCGAAAAGCTCTTCTGTTATAGTTTATATTCCAGCGATGTGGCCGGAGGAGCGGCAAAAACTCAAGAAAACCACTAAAGAGTAG
Protein
MAFEQQSAAAMQALQHQLLRGLSGAGGSGGMSAAAAMMQFTPLLYSYQLAMAQASALGKRYSMMEESTNPEESSNAITIQVDVHHRHYSQLSASIATTSHDCVNEPETTSDLPSIHRDYPKTAAQRMAEYRARKKKETPIVNHRKHKKSDAERAKEYRARKKAKIQSADAERAKEYRARNKAQIQEHTSQSQNFVESTSIQQSEHETNHLAYTDFHRHHLAHDEFQKKFVRNSFGYVCSVCERLWFKDDLRSASLQHNAILKTIVPHLDIKDVALCNTCVASLNNNSIPVMASYNGFKFPKKPDYLPPLDLVSERLISPRIPFMQIRRLRHVNGQYGIYGQIINVPVSIDSMVRTLPRSLSDDHCINVHIKRKKIHKSSYLHGIINKGTIKIWLQFLLKSPLYTMYDIKIDESFFENNDIDTIPQDEISEHVPIEESLVSQQQTLMWNEEKYLRLAPGEKNLPHSLLFDEHAEELSFPAIYLGQFRTFRDGVRVTPYTMATSELRRSDRRAVTPHHVLYVAMKIMRLRVRDSLTVAFKHIGKDTKITREQIENEDYVNNCIETNFAFLKSIPNSLWYWADKKKNLFAMIRQLGKPTIFLTISANEIGWTDLLQLLYKLKNNGAELSKKDAEELHYLEKVTLINEDSVTCAIYFNKLVNVLLNILQSPKKSPFGKYYVMHYFKRIEFQHRGSPHAHTLLWLANAPNDALGENKMDAISLINHLISISSKEASEHIKLQTHKHSFTCYKRISAHRPQQCRFEAPFLPSRSTEILLPIQQDEIGFNRYLQSYRNIRINLENNDYEDFDSFYQQNNISSDDEYRNILRAGIKRPRVFIKREPSEKWHNPFNPFILNLVKSNMDIQFITEEYSCAQYVAEYVNKTNRGISNLQREIIKCIDEHPEFDVVEITRKLGVEMLNSVEIPNQEAAWYLLREPMSKSSSVIVYIPAMWPEERQKLKKTTKE
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Cofactor
Mg(2+)
Similarity
Belongs to the helicase family.
Feature
chain ATP-dependent DNA helicase
Uniprot
H9JWT3
J9K9R0
J9JYX1
J9M7J3
H9J7F3
H9J482
+ More
A0A0C9Q9F9 A0A212FGN1 B0WBF3 B0WS82 B0X370 B0X2H4 B0WHR0 B0WQS5 A0A2W1BWM2 B0X8F9 B0W7Q6 B0XKN7 B0X3H8 B0W9W4 B0XE87 B0XBP3 J9M8Z2 B0XLK1 B0XHK1 B0WJB6 H9IWZ3 A0A1Y1NKE2 A0A2H9T4X5 S2JVK3 A0A0B7NNE4 A0A226D8L9 A0A226F1P1 A0A226EB64 A0A226F5N4 A0A0B7NFP9 A0A226DVC3 A0A163J913 A0A226ED60 A0A1C7MZM0 A0A0B7MTX6 A0A226DNF6 B0WH72 W4X9N8 W4YZ12 W4Z5F3 A0A1C7MYM4 A0A3B3CMZ8 A0A1A8U683 A0A1A8AX16 A0A1A8UT69 G3NDK2 A0A146TGF8 A0A146RI12 A0A146RH25
A0A0C9Q9F9 A0A212FGN1 B0WBF3 B0WS82 B0X370 B0X2H4 B0WHR0 B0WQS5 A0A2W1BWM2 B0X8F9 B0W7Q6 B0XKN7 B0X3H8 B0W9W4 B0XE87 B0XBP3 J9M8Z2 B0XLK1 B0XHK1 B0WJB6 H9IWZ3 A0A1Y1NKE2 A0A2H9T4X5 S2JVK3 A0A0B7NNE4 A0A226D8L9 A0A226F1P1 A0A226EB64 A0A226F5N4 A0A0B7NFP9 A0A226DVC3 A0A163J913 A0A226ED60 A0A1C7MZM0 A0A0B7MTX6 A0A226DNF6 B0WH72 W4X9N8 W4YZ12 W4Z5F3 A0A1C7MYM4 A0A3B3CMZ8 A0A1A8U683 A0A1A8AX16 A0A1A8UT69 G3NDK2 A0A146TGF8 A0A146RI12 A0A146RH25
EC Number
3.6.4.12
EMBL
BABH01018610
BABH01018611
BABH01018612
BABH01018613
BABH01018614
ABLF02018835
+ More
ABLF02018837 ABLF02018838 ABLF02009660 ABLF02060474 ABLF02010848 ABLF02010851 ABLF02010852 ABLF02024887 BABH01013754 BABH01032532 BABH01032533 BABH01032534 GBYB01011013 JAG80780.1 AGBW02008637 OWR52892.1 DS231879 EDS42392.1 DS232066 EDS33709.1 DS232310 EDS39572.1 DS232289 EDS39197.1 DS231939 EDS27927.1 DS232044 EDS32999.1 KZ149968 PZC76143.1 DS232488 EDS42495.1 DS231855 EDS38200.1 DS233927 EDS32356.1 DS232315 EDS39870.1 DS231866 EDS40531.1 DS232811 EDS45920.1 DS232646 EDS44392.1 ABLF02010071 DS234515 EDS34328.1 DS233163 EDS28452.1 DS231958 EDS29051.1 BABH01021151 BABH01021152 GEZM01005450 JAV96167.1 NSIT01000220 PJE78258.1 KE123980 EPB86818.1 LN733547 CEP17045.1 LNIX01000030 OXA41097.1 LNIX01000001 OXA63699.1 LNIX01000005 OXA54773.1 OXA64511.1 LN732935 CEP16237.1 LNIX01000011 OXA48651.1 LT553140 SAM00100.1 LNIX01000004 OXA55350.1 LUGH01000897 OBZ82310.1 LN719792 CEP08567.1 LNIX01000016 OXA46187.1 DS231933 EDS27530.1 AAGJ04118364 AAGJ04092821 AAGJ04070883 LUGH01001176 OBZ81506.1 HAEJ01003372 SBS43829.1 HADY01021084 SBP59569.1 HAEJ01010808 SBS51265.1 GCES01094404 JAQ91918.1 GCES01118596 JAQ67726.1 GCES01118597 JAQ67725.1
ABLF02018837 ABLF02018838 ABLF02009660 ABLF02060474 ABLF02010848 ABLF02010851 ABLF02010852 ABLF02024887 BABH01013754 BABH01032532 BABH01032533 BABH01032534 GBYB01011013 JAG80780.1 AGBW02008637 OWR52892.1 DS231879 EDS42392.1 DS232066 EDS33709.1 DS232310 EDS39572.1 DS232289 EDS39197.1 DS231939 EDS27927.1 DS232044 EDS32999.1 KZ149968 PZC76143.1 DS232488 EDS42495.1 DS231855 EDS38200.1 DS233927 EDS32356.1 DS232315 EDS39870.1 DS231866 EDS40531.1 DS232811 EDS45920.1 DS232646 EDS44392.1 ABLF02010071 DS234515 EDS34328.1 DS233163 EDS28452.1 DS231958 EDS29051.1 BABH01021151 BABH01021152 GEZM01005450 JAV96167.1 NSIT01000220 PJE78258.1 KE123980 EPB86818.1 LN733547 CEP17045.1 LNIX01000030 OXA41097.1 LNIX01000001 OXA63699.1 LNIX01000005 OXA54773.1 OXA64511.1 LN732935 CEP16237.1 LNIX01000011 OXA48651.1 LT553140 SAM00100.1 LNIX01000004 OXA55350.1 LUGH01000897 OBZ82310.1 LN719792 CEP08567.1 LNIX01000016 OXA46187.1 DS231933 EDS27530.1 AAGJ04118364 AAGJ04092821 AAGJ04070883 LUGH01001176 OBZ81506.1 HAEJ01003372 SBS43829.1 HADY01021084 SBP59569.1 HAEJ01010808 SBS51265.1 GCES01094404 JAQ91918.1 GCES01118596 JAQ67726.1 GCES01118597 JAQ67725.1
Proteomes
Pfam
PF05970 PIF1
+ More
PF14214 Helitron_like_N
PF16563 P66_CC
PF13359 DDE_Tnp_4
PF18358 Tudor_4
PF18359 TUDOR_5
PF02689 Herpes_Helicase
PF13538 UvrD_C_2
PF14529 Exo_endo_phos_2
PF02902 Peptidase_C48
PF02801 Ketoacyl-synt_C
PF00443 UCH
PF08659 KR
PF14765 PS-DH
PF00109 ketoacyl-synt
PF16197 KAsynt_C_assoc
PF00107 ADH_zinc_N
PF00698 Acyl_transf_1
PF00540 Gag_p17
PF02338 OTU
PF04843 Herpes_teg_N
PF03372 Exo_endo_phos
PF14214 Helitron_like_N
PF16563 P66_CC
PF13359 DDE_Tnp_4
PF18358 Tudor_4
PF18359 TUDOR_5
PF02689 Herpes_Helicase
PF13538 UvrD_C_2
PF14529 Exo_endo_phos_2
PF02902 Peptidase_C48
PF02801 Ketoacyl-synt_C
PF00443 UCH
PF08659 KR
PF14765 PS-DH
PF00109 ketoacyl-synt
PF16197 KAsynt_C_assoc
PF00107 ADH_zinc_N
PF00698 Acyl_transf_1
PF00540 Gag_p17
PF02338 OTU
PF04843 Herpes_teg_N
PF03372 Exo_endo_phos
Interpro
IPR010285
DNA_helicase_pif1-like
+ More
IPR025476 Helitron_helicase-like
IPR027417 P-loop_NTPase
IPR032346 P66_CC
IPR003593 AAA+_ATPase
IPR036691 Endo/exonu/phosph_ase_sf
IPR027806 HARBI1_dom
IPR003323 OTU_dom
IPR041292 Tudor_4
IPR041291 TUDOR_5
IPR009072 Histone-fold
IPR000558 Histone_H2B
IPR003840 DNA_helicase
IPR000014 PAS
IPR035965 PAS-like_dom_sf
IPR013087 Znf_C2H2_type
IPR027785 UvrD-like_helicase_C
IPR005135 Endo/exonuclease/phosphatase
IPR003653 Peptidase_C48_C
IPR038765 Papain-like_cys_pep_sf
IPR028889 USP_dom
IPR016039 Thiolase-like
IPR001394 Peptidase_C19_UCH
IPR014030 Ketoacyl_synth_N
IPR020843 PKS_ER
IPR013968 PKS_KR
IPR002364 Quin_OxRdtase/zeta-crystal_CS
IPR020801 PKS_acyl_transferase
IPR029063 SAM-dependent_MTases
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR001227 Ac_transferase_dom_sf
IPR020807 PKS_dehydratase
IPR036736 ACP-like_sf
IPR014031 Ketoacyl_synth_C
IPR016035 Acyl_Trfase/lysoPLipase
IPR013149 ADH_C
IPR032821 KAsynt_C_assoc
IPR011032 GroES-like_sf
IPR042104 PKS_dehydratase_sf
IPR014043 Acyl_transferase
IPR036291 NAD(P)-bd_dom_sf
IPR000071 Lentvrl_matrix_N
IPR011011 Znf_FYVE_PHD
IPR006928 Herpes_teg_USP
IPR019787 Znf_PHD-finger
IPR003410 HYR_dom
IPR036514 SGNH_hydro_sf
IPR025476 Helitron_helicase-like
IPR027417 P-loop_NTPase
IPR032346 P66_CC
IPR003593 AAA+_ATPase
IPR036691 Endo/exonu/phosph_ase_sf
IPR027806 HARBI1_dom
IPR003323 OTU_dom
IPR041292 Tudor_4
IPR041291 TUDOR_5
IPR009072 Histone-fold
IPR000558 Histone_H2B
IPR003840 DNA_helicase
IPR000014 PAS
IPR035965 PAS-like_dom_sf
IPR013087 Znf_C2H2_type
IPR027785 UvrD-like_helicase_C
IPR005135 Endo/exonuclease/phosphatase
IPR003653 Peptidase_C48_C
IPR038765 Papain-like_cys_pep_sf
IPR028889 USP_dom
IPR016039 Thiolase-like
IPR001394 Peptidase_C19_UCH
IPR014030 Ketoacyl_synth_N
IPR020843 PKS_ER
IPR013968 PKS_KR
IPR002364 Quin_OxRdtase/zeta-crystal_CS
IPR020801 PKS_acyl_transferase
IPR029063 SAM-dependent_MTases
IPR020841 PKS_Beta-ketoAc_synthase_dom
IPR001227 Ac_transferase_dom_sf
IPR020807 PKS_dehydratase
IPR036736 ACP-like_sf
IPR014031 Ketoacyl_synth_C
IPR016035 Acyl_Trfase/lysoPLipase
IPR013149 ADH_C
IPR032821 KAsynt_C_assoc
IPR011032 GroES-like_sf
IPR042104 PKS_dehydratase_sf
IPR014043 Acyl_transferase
IPR036291 NAD(P)-bd_dom_sf
IPR000071 Lentvrl_matrix_N
IPR011011 Znf_FYVE_PHD
IPR006928 Herpes_teg_USP
IPR019787 Znf_PHD-finger
IPR003410 HYR_dom
IPR036514 SGNH_hydro_sf
SUPFAM
CDD
ProteinModelPortal
H9JWT3
J9K9R0
J9JYX1
J9M7J3
H9J7F3
H9J482
+ More
A0A0C9Q9F9 A0A212FGN1 B0WBF3 B0WS82 B0X370 B0X2H4 B0WHR0 B0WQS5 A0A2W1BWM2 B0X8F9 B0W7Q6 B0XKN7 B0X3H8 B0W9W4 B0XE87 B0XBP3 J9M8Z2 B0XLK1 B0XHK1 B0WJB6 H9IWZ3 A0A1Y1NKE2 A0A2H9T4X5 S2JVK3 A0A0B7NNE4 A0A226D8L9 A0A226F1P1 A0A226EB64 A0A226F5N4 A0A0B7NFP9 A0A226DVC3 A0A163J913 A0A226ED60 A0A1C7MZM0 A0A0B7MTX6 A0A226DNF6 B0WH72 W4X9N8 W4YZ12 W4Z5F3 A0A1C7MYM4 A0A3B3CMZ8 A0A1A8U683 A0A1A8AX16 A0A1A8UT69 G3NDK2 A0A146TGF8 A0A146RI12 A0A146RH25
A0A0C9Q9F9 A0A212FGN1 B0WBF3 B0WS82 B0X370 B0X2H4 B0WHR0 B0WQS5 A0A2W1BWM2 B0X8F9 B0W7Q6 B0XKN7 B0X3H8 B0W9W4 B0XE87 B0XBP3 J9M8Z2 B0XLK1 B0XHK1 B0WJB6 H9IWZ3 A0A1Y1NKE2 A0A2H9T4X5 S2JVK3 A0A0B7NNE4 A0A226D8L9 A0A226F1P1 A0A226EB64 A0A226F5N4 A0A0B7NFP9 A0A226DVC3 A0A163J913 A0A226ED60 A0A1C7MZM0 A0A0B7MTX6 A0A226DNF6 B0WH72 W4X9N8 W4YZ12 W4Z5F3 A0A1C7MYM4 A0A3B3CMZ8 A0A1A8U683 A0A1A8AX16 A0A1A8UT69 G3NDK2 A0A146TGF8 A0A146RI12 A0A146RH25
Ontologies
GO
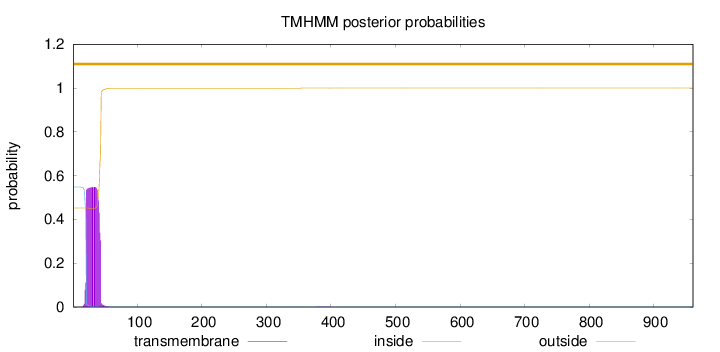
Topology
Length:
961
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.1567699999999
Exp number, first 60 AAs:
12.14547
Total prob of N-in:
0.54839
POSSIBLE N-term signal
sequence
outside
1 - 961
Population Genetic Test Statistics
Pi
206.72602
Theta
195.783349
Tajima's D
0.217015
CLR
18.916321
CSRT
0.435578221088946
Interpretation
Uncertain
