Gene
KWMTBOMO16263
Annotation
hypothetical_protein_RF55_15411_[Lasius_niger]
Location in the cell
Nuclear Reliability : 3.728
Sequence
CDS
ATGTTATCGTGTTTTTTTTCAGGATTTTTCCAATTTAATGAGCAACGTGTCAGGCAACTTGTCAGTGATGTTGAGTTGGGTGACCGCCGTCCTTCGCAGTTTTTACGTTATCTTAGATCATTGGCAGGAAAAACCCTCACTGATGAAAATTTGCTTCGCCAGCTATGGATGCGTCGCCTCCCACAACATCTCCAAGCAATTTTGGCCGCCCAATCAGAACTCTCGCTGGACAAAGTCGCTGAGCTAGCGGACAAAATTTTGGAAGTTTCGCCTGGTATGGTCGCACCTCATTCATCTGCTGTTTTCTCAGCAAGTACGTCTGCCGTCTCAACCCCTTCGTTAGAAGATATCACACGGCGTCTGGATGAGGTCACGCGTCAGCTTGCTGTGCTTACTGCAGAGCGCGGTAGAAATAGGACCCGGGATTTCTCAAACCAGCGACGACGCTCACAAACGCCTGGTAAGCTTCAAATGTGCTTTTACCATAGGAAATTTAATACTAAAGCTGTTAAGTGTATTCAACCATGTACCTGGACGCCGCCGCCATCTTCGGAAAACGACAAAAGCAGTCAATGA
Protein
MLSCFFSGFFQFNEQRVRQLVSDVELGDRRPSQFLRYLRSLAGKTLTDENLLRQLWMRRLPQHLQAILAAQSELSLDKVAELADKILEVSPGMVAPHSSAVFSASTSAVSTPSLEDITRRLDEVTRQLAVLTAERGRNRTRDFSNQRRRSQTPGKLQMCFYHRKFNTKAVKCIQPCTWTPPPSSENDKSSQ
Summary
Uniprot
A0A1E1W1E7
A0A0J7K6I6
A0A0J7K6V3
A0A085NBV3
A0A151JCH0
A0A151ITG2
+ More
A0A0J7K7T5 A0A151IZ47 T1IE13 A0A151J3U8 A0A0L7QJB5 A0A1Y1MA44 A0A1W4XBP5 A0A085LTM6 A0A0J7K6Y3 A0A085N4U2 A0A2L2Z3N6 A0A154P674 A0A026VRX8 A0A0L7QK36 A0A1E1X2S5 E2B0A9 A0A0J7KD10 A0A0J7K6V1 A0A1Y1MXV9 A0A151IRN4 A0A0J7JWP5 A0A154PA69 A0A2A4K2H4 T1I831 A0A2J7PYD5 A0A2H1WXE7 A0A094ZSI3 A0A0L7QJ97 A0A151JF00 C7C1Z8 A0A2G8L3A3 A0A151K1K5 A0A0L0ULM5 A0A3P7ZWB0 G4M0C7 A0A0J7NAK0 A0A183NPV5 A0A183NQG5 A0A0J7K0A7 A0A2J7RQK2 G4LW71 A0A3L8DMT5 A0A2A4JHF4 A0A095A584 A0A224XWW2 A0A131YTY1 C4QHY4 A0A0K8SFH9 A0A154PMS5 A0A151K0A8 D6WTN9 G4LWP6 A0A1W4X916 G4VT83 G4LVX0 G4VLV9 T1HU96 A0A2J7QTC8 A0A2J7PLR6 A0A2A4J8M7 G4VSL1 A0A139W8G7 A0A0L7LFW5 G4VH89 D7GYF3 G4VI25 G4VMA7 D7EJX5 G4M134 A0A0K8SV59 A0A131YM78 A0A0P8Y533 G4VRT0 A0A139WA14 A0A131Z7F2 G4V794 A0A3Q0KTC5 G4V682 A0A3Q0KKC0 A0A095C1W4 G4VET2 G4V8M5 G4VR49 C4Q8R4 A0A026WEP3 A0A183B8I5 C1M100 C1M2Q9 A0A0L7QJ51 G4LZM4 G4VLM4 C1M2I0
A0A0J7K7T5 A0A151IZ47 T1IE13 A0A151J3U8 A0A0L7QJB5 A0A1Y1MA44 A0A1W4XBP5 A0A085LTM6 A0A0J7K6Y3 A0A085N4U2 A0A2L2Z3N6 A0A154P674 A0A026VRX8 A0A0L7QK36 A0A1E1X2S5 E2B0A9 A0A0J7KD10 A0A0J7K6V1 A0A1Y1MXV9 A0A151IRN4 A0A0J7JWP5 A0A154PA69 A0A2A4K2H4 T1I831 A0A2J7PYD5 A0A2H1WXE7 A0A094ZSI3 A0A0L7QJ97 A0A151JF00 C7C1Z8 A0A2G8L3A3 A0A151K1K5 A0A0L0ULM5 A0A3P7ZWB0 G4M0C7 A0A0J7NAK0 A0A183NPV5 A0A183NQG5 A0A0J7K0A7 A0A2J7RQK2 G4LW71 A0A3L8DMT5 A0A2A4JHF4 A0A095A584 A0A224XWW2 A0A131YTY1 C4QHY4 A0A0K8SFH9 A0A154PMS5 A0A151K0A8 D6WTN9 G4LWP6 A0A1W4X916 G4VT83 G4LVX0 G4VLV9 T1HU96 A0A2J7QTC8 A0A2J7PLR6 A0A2A4J8M7 G4VSL1 A0A139W8G7 A0A0L7LFW5 G4VH89 D7GYF3 G4VI25 G4VMA7 D7EJX5 G4M134 A0A0K8SV59 A0A131YM78 A0A0P8Y533 G4VRT0 A0A139WA14 A0A131Z7F2 G4V794 A0A3Q0KTC5 G4V682 A0A3Q0KKC0 A0A095C1W4 G4VET2 G4V8M5 G4VR49 C4Q8R4 A0A026WEP3 A0A183B8I5 C1M100 C1M2Q9 A0A0L7QJ51 G4LZM4 G4VLM4 C1M2I0
Pubmed
EMBL
GDQN01010239
JAT80815.1
LBMM01013103
KMQ85816.1
LBMM01012479
KMQ86203.1
+ More
KL367519 KFD66949.1 KQ979048 KYN23218.1 KQ981018 KYN10257.1 LBMM01012075 KMQ86422.1 KQ980726 KYN14011.1 ACPB03037918 KQ980247 KYN17109.1 LHQN01021927 LHQN01025379 KOC58628.1 KOC58659.1 GEZM01036621 JAV82564.1 KL363297 KFD48322.1 LBMM01012610 KMQ86112.1 KL367555 KFD64488.1 IAAA01082076 LAA14320.1 KQ434816 KZC06854.1 KK111495 EZA46390.1 KQ414987 KOC58988.1 GFAC01005832 JAT93356.1 GL444488 EFN60880.1 LBMM01009142 KMQ88358.1 LBMM01012923 LBMM01012917 KMQ85926.1 KMQ85930.1 GEZM01017686 JAV90494.1 KQ981124 KYN09337.1 LBMM01026224 KMQ82336.1 KQ434840 KZC08080.1 NWSH01000267 PCG77880.1 ACPB03012735 NEVH01020366 PNF21359.1 ODYU01011787 SOQ57731.1 KL250741 KGB36059.1 KQ415240 KOC58703.1 KQ979024 KYN24351.1 FN356212 CAX83700.1 MRZV01001127 MRZV01000236 PIK40395.1 PIK54731.1 LKEX01008659 KYN50015.1 AJIL01003693 KNE87923.1 UZAL01011781 VDP05747.1 CABG01000076 CCD60595.1 LBMM01007548 KMQ89630.1 UZAL01010187 VDP03243.1 LBMM01018089 KMQ83878.1 NEVH01000707 PNF43124.1 CABG01000023 CCD59159.1 QOIP01000006 RLU21744.1 NWSH01001575 PCG70822.1 KL252356 KGB42042.1 GFTR01003955 JAW12471.1 GEDV01006140 JAP82417.1 CABG01000346 CAZ35068.1 GBRD01013927 JAG51899.1 KQ434992 KZC13185.1 KQ981301 KYN43041.1 KQ971355 EFA07205.2 CABG01000031 CCD59329.1 HE601631 CCD83017.1 CABG01000020 CCD59058.1 HE601629 CCD80464.1 ACPB03016217 NEVH01011195 PNF31836.1 NEVH01024424 PNF17276.1 NWSH01002598 PCG67884.1 CCD81852.1 KQ973343 KXZ75568.1 JTDY01001300 KOB74269.1 HE601627 CCD79752.1 GG695519 EFA13383.1 CCD79076.1 CCD80612.1 KQ971759 EFA12902.2 CABG01000094 CCD60852.1 GBRD01008790 JAG57031.1 GEDV01008183 JAP80374.1 CH905996 KPU81830.1 CCD82772.1 KQ971745 KYB24749.1 GEDV01001795 JAP86762.1 HE601624 CCD74881.1 CCD74711.1 KL250698 KGB35593.1 HE601626 CCD78440.1 CCD75122.1 CCD81588.1 HE601625 CCD77013.1 CCD77288.1 CCD80388.1 CCD80486.1 CCD81882.1 KK107241 EZA54570.1 UZAN01060843 VDP92792.1 HE601630 CCD75315.1 CCD80887.1 CCD75326.1 CCD79184.1 CCD79457.1 LHQN01026442 KQ414593 KOC58672.1 KOC70162.1 CABG01000063 CCD60342.1 CCD80379.1 CABG01000118 CAY18932.2
KL367519 KFD66949.1 KQ979048 KYN23218.1 KQ981018 KYN10257.1 LBMM01012075 KMQ86422.1 KQ980726 KYN14011.1 ACPB03037918 KQ980247 KYN17109.1 LHQN01021927 LHQN01025379 KOC58628.1 KOC58659.1 GEZM01036621 JAV82564.1 KL363297 KFD48322.1 LBMM01012610 KMQ86112.1 KL367555 KFD64488.1 IAAA01082076 LAA14320.1 KQ434816 KZC06854.1 KK111495 EZA46390.1 KQ414987 KOC58988.1 GFAC01005832 JAT93356.1 GL444488 EFN60880.1 LBMM01009142 KMQ88358.1 LBMM01012923 LBMM01012917 KMQ85926.1 KMQ85930.1 GEZM01017686 JAV90494.1 KQ981124 KYN09337.1 LBMM01026224 KMQ82336.1 KQ434840 KZC08080.1 NWSH01000267 PCG77880.1 ACPB03012735 NEVH01020366 PNF21359.1 ODYU01011787 SOQ57731.1 KL250741 KGB36059.1 KQ415240 KOC58703.1 KQ979024 KYN24351.1 FN356212 CAX83700.1 MRZV01001127 MRZV01000236 PIK40395.1 PIK54731.1 LKEX01008659 KYN50015.1 AJIL01003693 KNE87923.1 UZAL01011781 VDP05747.1 CABG01000076 CCD60595.1 LBMM01007548 KMQ89630.1 UZAL01010187 VDP03243.1 LBMM01018089 KMQ83878.1 NEVH01000707 PNF43124.1 CABG01000023 CCD59159.1 QOIP01000006 RLU21744.1 NWSH01001575 PCG70822.1 KL252356 KGB42042.1 GFTR01003955 JAW12471.1 GEDV01006140 JAP82417.1 CABG01000346 CAZ35068.1 GBRD01013927 JAG51899.1 KQ434992 KZC13185.1 KQ981301 KYN43041.1 KQ971355 EFA07205.2 CABG01000031 CCD59329.1 HE601631 CCD83017.1 CABG01000020 CCD59058.1 HE601629 CCD80464.1 ACPB03016217 NEVH01011195 PNF31836.1 NEVH01024424 PNF17276.1 NWSH01002598 PCG67884.1 CCD81852.1 KQ973343 KXZ75568.1 JTDY01001300 KOB74269.1 HE601627 CCD79752.1 GG695519 EFA13383.1 CCD79076.1 CCD80612.1 KQ971759 EFA12902.2 CABG01000094 CCD60852.1 GBRD01008790 JAG57031.1 GEDV01008183 JAP80374.1 CH905996 KPU81830.1 CCD82772.1 KQ971745 KYB24749.1 GEDV01001795 JAP86762.1 HE601624 CCD74881.1 CCD74711.1 KL250698 KGB35593.1 HE601626 CCD78440.1 CCD75122.1 CCD81588.1 HE601625 CCD77013.1 CCD77288.1 CCD80388.1 CCD80486.1 CCD81882.1 KK107241 EZA54570.1 UZAN01060843 VDP92792.1 HE601630 CCD75315.1 CCD80887.1 CCD75326.1 CCD79184.1 CCD79457.1 LHQN01026442 KQ414593 KOC58672.1 KOC70162.1 CABG01000063 CCD60342.1 CCD80379.1 CABG01000118 CAY18932.2
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A1E1W1E7
A0A0J7K6I6
A0A0J7K6V3
A0A085NBV3
A0A151JCH0
A0A151ITG2
+ More
A0A0J7K7T5 A0A151IZ47 T1IE13 A0A151J3U8 A0A0L7QJB5 A0A1Y1MA44 A0A1W4XBP5 A0A085LTM6 A0A0J7K6Y3 A0A085N4U2 A0A2L2Z3N6 A0A154P674 A0A026VRX8 A0A0L7QK36 A0A1E1X2S5 E2B0A9 A0A0J7KD10 A0A0J7K6V1 A0A1Y1MXV9 A0A151IRN4 A0A0J7JWP5 A0A154PA69 A0A2A4K2H4 T1I831 A0A2J7PYD5 A0A2H1WXE7 A0A094ZSI3 A0A0L7QJ97 A0A151JF00 C7C1Z8 A0A2G8L3A3 A0A151K1K5 A0A0L0ULM5 A0A3P7ZWB0 G4M0C7 A0A0J7NAK0 A0A183NPV5 A0A183NQG5 A0A0J7K0A7 A0A2J7RQK2 G4LW71 A0A3L8DMT5 A0A2A4JHF4 A0A095A584 A0A224XWW2 A0A131YTY1 C4QHY4 A0A0K8SFH9 A0A154PMS5 A0A151K0A8 D6WTN9 G4LWP6 A0A1W4X916 G4VT83 G4LVX0 G4VLV9 T1HU96 A0A2J7QTC8 A0A2J7PLR6 A0A2A4J8M7 G4VSL1 A0A139W8G7 A0A0L7LFW5 G4VH89 D7GYF3 G4VI25 G4VMA7 D7EJX5 G4M134 A0A0K8SV59 A0A131YM78 A0A0P8Y533 G4VRT0 A0A139WA14 A0A131Z7F2 G4V794 A0A3Q0KTC5 G4V682 A0A3Q0KKC0 A0A095C1W4 G4VET2 G4V8M5 G4VR49 C4Q8R4 A0A026WEP3 A0A183B8I5 C1M100 C1M2Q9 A0A0L7QJ51 G4LZM4 G4VLM4 C1M2I0
A0A0J7K7T5 A0A151IZ47 T1IE13 A0A151J3U8 A0A0L7QJB5 A0A1Y1MA44 A0A1W4XBP5 A0A085LTM6 A0A0J7K6Y3 A0A085N4U2 A0A2L2Z3N6 A0A154P674 A0A026VRX8 A0A0L7QK36 A0A1E1X2S5 E2B0A9 A0A0J7KD10 A0A0J7K6V1 A0A1Y1MXV9 A0A151IRN4 A0A0J7JWP5 A0A154PA69 A0A2A4K2H4 T1I831 A0A2J7PYD5 A0A2H1WXE7 A0A094ZSI3 A0A0L7QJ97 A0A151JF00 C7C1Z8 A0A2G8L3A3 A0A151K1K5 A0A0L0ULM5 A0A3P7ZWB0 G4M0C7 A0A0J7NAK0 A0A183NPV5 A0A183NQG5 A0A0J7K0A7 A0A2J7RQK2 G4LW71 A0A3L8DMT5 A0A2A4JHF4 A0A095A584 A0A224XWW2 A0A131YTY1 C4QHY4 A0A0K8SFH9 A0A154PMS5 A0A151K0A8 D6WTN9 G4LWP6 A0A1W4X916 G4VT83 G4LVX0 G4VLV9 T1HU96 A0A2J7QTC8 A0A2J7PLR6 A0A2A4J8M7 G4VSL1 A0A139W8G7 A0A0L7LFW5 G4VH89 D7GYF3 G4VI25 G4VMA7 D7EJX5 G4M134 A0A0K8SV59 A0A131YM78 A0A0P8Y533 G4VRT0 A0A139WA14 A0A131Z7F2 G4V794 A0A3Q0KTC5 G4V682 A0A3Q0KKC0 A0A095C1W4 G4VET2 G4V8M5 G4VR49 C4Q8R4 A0A026WEP3 A0A183B8I5 C1M100 C1M2Q9 A0A0L7QJ51 G4LZM4 G4VLM4 C1M2I0
Ontologies
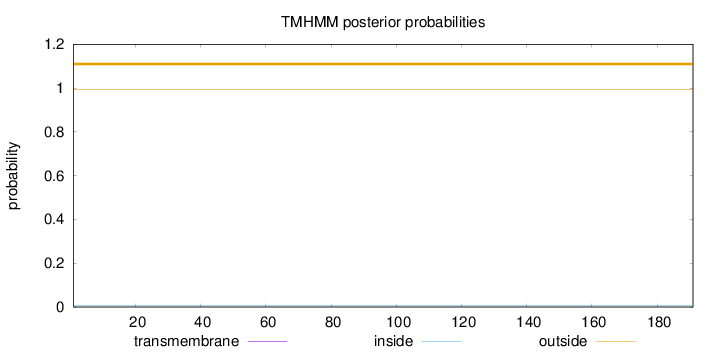
Topology
Length:
191
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00081
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00543
outside
1 - 191
Population Genetic Test Statistics
Pi
335.302981
Theta
176.121181
Tajima's D
2.863764
CLR
0
CSRT
0.972951352432378
Interpretation
Uncertain
