Gene
KWMTBOMO16258 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013898
Annotation
proteasome_zeta_subunit_[Bombyx_mori]
Full name
Proteasome endopeptidase complex
+ More
Proteasome subunit alpha type
Proteasome subunit alpha type
Location in the cell
Cytoplasmic Reliability : 2.231
Sequence
CDS
ATGTTTCTCACACGTTCCGAGTATGATCGCGGCGTCAACACATTCAGCCCTGAAGGAAGGCTGTTCCAGGTGGAATACGCCATAGAAGCTATCAAACTTGGCTCCACTGCAATTGGCATTTGTACTTCGGAGGGAGTTGTTTTGGCGGTGGAAAAAAGGATAACTTCAACTTTGATGGAACCCACGACCATCGAAAAGATTGTTGAAGTAGATCGACACATCGCTTGCGCCGTTTCGGGTCTAATGGCCGACTCCAGGACCTTAGTTGAAAGGGCCCGAGTTGAATGTCAAAACCACTGGTTTGTGTACAATGAGCGTATGAGTGTAGAATCATGTGCTCAGGCCGTTTCCAACTTAGCTATACAGTTTGGAGACAGTGACGATGATAGCGGAACAGCCATGTCACGGCCATTTGGTGTTGCTGTTATGTTTGCAGGCATTGATGAGAAAGGCCCTCAACTGTTTCATATGGACCCGAGTGGTACATTTGTGCAGTATGATGCCAAGGCCATTGGTTCTGGCAGTGAAGGTGCCCAGCAGAGTTTGAAGGAGATTTATCACAAGTCGATGACCCTCAAAGAGGCCATCAAGTCTGCACTCACCATACTGAAGCAGGTCATGGAGGAAAAGCTCTCGGAGAACAATGTTGAAGTAGTGACAATGACTCCTGGATCATTGTTCCACATGTTCACAAGGGAACAACTTGCTGAGGTTATCAAAGACATACCTTAA
Protein
MFLTRSEYDRGVNTFSPEGRLFQVEYAIEAIKLGSTAIGICTSEGVVLAVEKRITSTLMEPTTIEKIVEVDRHIACAVSGLMADSRTLVERARVECQNHWFVYNERMSVESCAQAVSNLAIQFGDSDDDSGTAMSRPFGVAVMFAGIDEKGPQLFHMDPSGTFVQYDAKAIGSGSEGAQQSLKEIYHKSMTLKEAIKSALTILKQVMEEKLSENNVEVVTMTPGSLFHMFTREQLAEVIKDIP
Summary
Description
The proteasome is a multicatalytic proteinase complex which is characterized by its ability to cleave peptides with Arg, Phe, Tyr, Leu, and Glu adjacent to the leaving group at neutral or slightly basic pH.
Catalytic Activity
Cleavage of peptide bonds with very broad specificity.
Subunit
The 20S proteasome core is composed of 28 subunits that are arranged in four stacked rings, resulting in a barrel-shaped structure. The two end rings are each formed by seven alpha subunits, and the two central rings are each formed by seven beta subunits.
The 26S proteasome consists of a 20S proteasome core and two 19S regulatory subunits.
The 26S proteasome consists of a 20S proteasome core and two 19S regulatory subunits.
Similarity
Belongs to the peptidase T1A family.
Uniprot
H9JWI4
Q2F661
A0A2A4JJ31
A0A2H1VS21
A0A1E1W968
S4PBH6
+ More
A0A2W1B5X0 A0A2A4JKH8 A0A194Q2V7 A0A212FER0 E9IFI5 A0A151IK79 A0A139WFD4 A0A1S6Q5L3 F4X885 A0A158NBQ2 D6WT31 E2BCA4 A0A026WC60 A0A0L7QU49 A0A2J7Q9G1 A0A2P8Z4B7 A0A067RTL6 A0A0M9A518 E2A1P1 A0A087ZZN8 A0A195FD54 A0A151X9I5 A0A195BG81 A0A195DS04 A0A1Y1N7Z0 A0A0J7K3A1 K7IUK2 J3JTS6 N6TD48 A0A1B6CPF7 A0A232F5X5 A0A0P5IE39 A0A154P3U7 E9HGR7 A0A0P5G816 A0A0P4ZDQ8 A0A0P6E054 A0A0P5G0X2 A0A0N8C4N2 A0A0P6FYK7 A0A0P6EKE3 A0A0P5S5S9 A0A0P5JKP0 A0A0P5ASF6 A0A1B6EX92 A0A0P5HLG0 A0A0P6DJ38 A0A1B6H9C8 A0A0P6GW73 A0A0P5VCU5 A0A0P5KUL8 A0A0P5NHZ6 A0A0P4XSD7 A0A1B6L3Y8 T1JK43 A0A0P5SI01 A0A0P5G1X0 A0A0K8RHF0 A0A0P5V5N8 A0A0P5NQ08 T1DEV5 A0A0P5A6A2 Q1HQN1 A0A182GLM4 A0A023GGP0 G3MMH6 A0A1E1X441 E0VDI1 A0A2R5LG22 A0A0P6G830 A0A293MYB5 A0A023FK49 A0A023FXX5 A0A3S3PEI0 A0A1S4JRU4 A0A0P5SHE8 A0A310SRT7 A0A131XDD5 A0A226EMH8 A0A1D2MQC3 A0A224YRP6 A0A131Z3A6 L7M5R1 V4AEP3 B0WTL4 U5EMA6 A0A023EN66 S4REA7 A0A1Z5KXE3 A0A1Q3FDE8 C3ZPP8 A0A0P5VYK8 Q3TUI9 A0A1S3GET8
A0A2W1B5X0 A0A2A4JKH8 A0A194Q2V7 A0A212FER0 E9IFI5 A0A151IK79 A0A139WFD4 A0A1S6Q5L3 F4X885 A0A158NBQ2 D6WT31 E2BCA4 A0A026WC60 A0A0L7QU49 A0A2J7Q9G1 A0A2P8Z4B7 A0A067RTL6 A0A0M9A518 E2A1P1 A0A087ZZN8 A0A195FD54 A0A151X9I5 A0A195BG81 A0A195DS04 A0A1Y1N7Z0 A0A0J7K3A1 K7IUK2 J3JTS6 N6TD48 A0A1B6CPF7 A0A232F5X5 A0A0P5IE39 A0A154P3U7 E9HGR7 A0A0P5G816 A0A0P4ZDQ8 A0A0P6E054 A0A0P5G0X2 A0A0N8C4N2 A0A0P6FYK7 A0A0P6EKE3 A0A0P5S5S9 A0A0P5JKP0 A0A0P5ASF6 A0A1B6EX92 A0A0P5HLG0 A0A0P6DJ38 A0A1B6H9C8 A0A0P6GW73 A0A0P5VCU5 A0A0P5KUL8 A0A0P5NHZ6 A0A0P4XSD7 A0A1B6L3Y8 T1JK43 A0A0P5SI01 A0A0P5G1X0 A0A0K8RHF0 A0A0P5V5N8 A0A0P5NQ08 T1DEV5 A0A0P5A6A2 Q1HQN1 A0A182GLM4 A0A023GGP0 G3MMH6 A0A1E1X441 E0VDI1 A0A2R5LG22 A0A0P6G830 A0A293MYB5 A0A023FK49 A0A023FXX5 A0A3S3PEI0 A0A1S4JRU4 A0A0P5SHE8 A0A310SRT7 A0A131XDD5 A0A226EMH8 A0A1D2MQC3 A0A224YRP6 A0A131Z3A6 L7M5R1 V4AEP3 B0WTL4 U5EMA6 A0A023EN66 S4REA7 A0A1Z5KXE3 A0A1Q3FDE8 C3ZPP8 A0A0P5VYK8 Q3TUI9 A0A1S3GET8
EC Number
3.4.25.1
Pubmed
19121390
23622113
28756777
26354079
22118469
21282665
+ More
18362917 19820115 21719571 21347285 20798317 24508170 30249741 29403074 24845553 28004739 20075255 22516182 23537049 28648823 21292972 24330624 17204158 17510324 26483478 22216098 28503490 20566863 28049606 27289101 28797301 26830274 25576852 23254933 24945155 28528879 18563158 10349636 11042159 11076861 11217851 12466851 16141073
18362917 19820115 21719571 21347285 20798317 24508170 30249741 29403074 24845553 28004739 20075255 22516182 23537049 28648823 21292972 24330624 17204158 17510324 26483478 22216098 28503490 20566863 28049606 27289101 28797301 26830274 25576852 23254933 24945155 28528879 18563158 10349636 11042159 11076861 11217851 12466851 16141073
EMBL
BABH01018608
DQ311211
ABD36156.1
NWSH01001212
PCG72105.1
ODYU01004100
+ More
SOQ43619.1 GDQN01007541 JAT83513.1 GAIX01004616 JAA87944.1 KZ150476 PZC70741.1 PCG72104.1 KQ459588 KPI97740.1 AGBW02008912 OWR52198.1 GL762841 EFZ20649.1 KQ977242 KYN04674.1 KQ971354 KYB26545.1 KY628452 AQV09455.1 GL888932 EGI57220.1 ADTU01011278 EFA05871.2 GL447257 EFN86640.1 KK107276 QOIP01000004 EZA53647.1 RLU24036.1 KQ414740 KOC62084.1 NEVH01016358 PNF25218.1 PYGN01000203 PSN51317.1 KK852471 KDR23179.1 KQ435756 KOX76009.1 GL435766 EFN72714.1 KQ981673 KYN38333.1 KQ982373 KYQ56980.1 KQ976500 KYM83214.1 KQ980581 KYN15299.1 GEZM01010547 GEZM01010546 JAV94001.1 LBMM01015431 KMQ84812.1 AAZX01007579 BT126635 AEE61598.1 APGK01035321 KB740923 KB632333 ENN78269.1 ERL92699.1 GEDC01022053 JAS15245.1 NNAY01000846 OXU26221.1 GDIQ01214655 JAK37070.1 KQ434809 KZC06615.1 GL732643 EFX69073.1 GDIQ01244959 GDIQ01242661 GDIQ01152233 GDIQ01107049 LRGB01000626 JAK06766.1 KZS17458.1 GDIP01215741 JAJ07661.1 GDIQ01084712 JAN10025.1 GDIQ01247808 JAK03917.1 GDIQ01115203 JAL36523.1 GDIQ01041277 JAN53460.1 GDIQ01061321 JAN33416.1 GDIQ01092550 JAL59176.1 GDIQ01197020 JAK54705.1 GDIP01196297 GDIP01195098 JAJ28304.1 GECZ01027170 JAS42599.1 GDIQ01231710 JAK20015.1 GDIQ01078213 JAN16524.1 GECU01036484 JAS71222.1 GDIQ01038275 JAN56462.1 GDIP01102105 JAM01610.1 GDIQ01179022 JAK72703.1 GDIQ01145842 JAL05884.1 GDIP01237247 JAI86154.1 GEBQ01021733 JAT18244.1 AFFK01019071 GDIP01139705 JAL64009.1 GDIQ01247809 JAK03916.1 GADI01003879 JAA69929.1 GDIP01105008 JAL98706.1 GDIP01232197 GDIQ01160921 JAI91204.1 JAK90804.1 GALA01000902 JAA93950.1 GDIP01207894 JAJ15508.1 DQ440413 CH477299 ABF18446.1 EAT44303.1 JXUM01168979 KQ577807 KXJ67791.1 GBBM01002296 JAC33122.1 JO843077 AEO34694.1 GFAC01005392 JAT93796.1 DS235078 EEB11437.1 GGLE01004350 MBY08476.1 GDIQ01047583 JAN47154.1 GFWV01021075 MAA45803.1 GBBK01003479 JAC21003.1 GBBL01001770 JAC25550.1 NCKU01015899 RWR99284.1 GDIP01139706 JAL64008.1 KQ761155 OAD58116.1 GEFH01003994 JAP64587.1 LNIX01000003 OXA58842.1 LJIJ01000693 ODM95267.1 GFPF01005498 MAA16644.1 GEDV01002673 JAP85884.1 GACK01005837 JAA59197.1 KB201656 ESO95327.1 DS232088 EDS34463.1 GANO01001115 JAB58756.1 GAPW01003784 JAC09814.1 GFJQ02007342 JAV99627.1 GFDL01009506 JAV25539.1 GG666659 EEN45469.1 GDIP01093363 JAM10352.1 AK160741 BAE35982.1
SOQ43619.1 GDQN01007541 JAT83513.1 GAIX01004616 JAA87944.1 KZ150476 PZC70741.1 PCG72104.1 KQ459588 KPI97740.1 AGBW02008912 OWR52198.1 GL762841 EFZ20649.1 KQ977242 KYN04674.1 KQ971354 KYB26545.1 KY628452 AQV09455.1 GL888932 EGI57220.1 ADTU01011278 EFA05871.2 GL447257 EFN86640.1 KK107276 QOIP01000004 EZA53647.1 RLU24036.1 KQ414740 KOC62084.1 NEVH01016358 PNF25218.1 PYGN01000203 PSN51317.1 KK852471 KDR23179.1 KQ435756 KOX76009.1 GL435766 EFN72714.1 KQ981673 KYN38333.1 KQ982373 KYQ56980.1 KQ976500 KYM83214.1 KQ980581 KYN15299.1 GEZM01010547 GEZM01010546 JAV94001.1 LBMM01015431 KMQ84812.1 AAZX01007579 BT126635 AEE61598.1 APGK01035321 KB740923 KB632333 ENN78269.1 ERL92699.1 GEDC01022053 JAS15245.1 NNAY01000846 OXU26221.1 GDIQ01214655 JAK37070.1 KQ434809 KZC06615.1 GL732643 EFX69073.1 GDIQ01244959 GDIQ01242661 GDIQ01152233 GDIQ01107049 LRGB01000626 JAK06766.1 KZS17458.1 GDIP01215741 JAJ07661.1 GDIQ01084712 JAN10025.1 GDIQ01247808 JAK03917.1 GDIQ01115203 JAL36523.1 GDIQ01041277 JAN53460.1 GDIQ01061321 JAN33416.1 GDIQ01092550 JAL59176.1 GDIQ01197020 JAK54705.1 GDIP01196297 GDIP01195098 JAJ28304.1 GECZ01027170 JAS42599.1 GDIQ01231710 JAK20015.1 GDIQ01078213 JAN16524.1 GECU01036484 JAS71222.1 GDIQ01038275 JAN56462.1 GDIP01102105 JAM01610.1 GDIQ01179022 JAK72703.1 GDIQ01145842 JAL05884.1 GDIP01237247 JAI86154.1 GEBQ01021733 JAT18244.1 AFFK01019071 GDIP01139705 JAL64009.1 GDIQ01247809 JAK03916.1 GADI01003879 JAA69929.1 GDIP01105008 JAL98706.1 GDIP01232197 GDIQ01160921 JAI91204.1 JAK90804.1 GALA01000902 JAA93950.1 GDIP01207894 JAJ15508.1 DQ440413 CH477299 ABF18446.1 EAT44303.1 JXUM01168979 KQ577807 KXJ67791.1 GBBM01002296 JAC33122.1 JO843077 AEO34694.1 GFAC01005392 JAT93796.1 DS235078 EEB11437.1 GGLE01004350 MBY08476.1 GDIQ01047583 JAN47154.1 GFWV01021075 MAA45803.1 GBBK01003479 JAC21003.1 GBBL01001770 JAC25550.1 NCKU01015899 RWR99284.1 GDIP01139706 JAL64008.1 KQ761155 OAD58116.1 GEFH01003994 JAP64587.1 LNIX01000003 OXA58842.1 LJIJ01000693 ODM95267.1 GFPF01005498 MAA16644.1 GEDV01002673 JAP85884.1 GACK01005837 JAA59197.1 KB201656 ESO95327.1 DS232088 EDS34463.1 GANO01001115 JAB58756.1 GAPW01003784 JAC09814.1 GFJQ02007342 JAV99627.1 GFDL01009506 JAV25539.1 GG666659 EEN45469.1 GDIP01093363 JAM10352.1 AK160741 BAE35982.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000078542
UP000007266
+ More
UP000007755 UP000005205 UP000008237 UP000053097 UP000279307 UP000053825 UP000235965 UP000245037 UP000027135 UP000053105 UP000000311 UP000005203 UP000078541 UP000075809 UP000078540 UP000078492 UP000036403 UP000002358 UP000019118 UP000030742 UP000215335 UP000076502 UP000000305 UP000076858 UP000008820 UP000069940 UP000249989 UP000009046 UP000285301 UP000198287 UP000094527 UP000030746 UP000002320 UP000245300 UP000001554 UP000081671
UP000007755 UP000005205 UP000008237 UP000053097 UP000279307 UP000053825 UP000235965 UP000245037 UP000027135 UP000053105 UP000000311 UP000005203 UP000078541 UP000075809 UP000078540 UP000078492 UP000036403 UP000002358 UP000019118 UP000030742 UP000215335 UP000076502 UP000000305 UP000076858 UP000008820 UP000069940 UP000249989 UP000009046 UP000285301 UP000198287 UP000094527 UP000030746 UP000002320 UP000245300 UP000001554 UP000081671
Interpro
SUPFAM
SSF56235
SSF56235
Gene 3D
ProteinModelPortal
H9JWI4
Q2F661
A0A2A4JJ31
A0A2H1VS21
A0A1E1W968
S4PBH6
+ More
A0A2W1B5X0 A0A2A4JKH8 A0A194Q2V7 A0A212FER0 E9IFI5 A0A151IK79 A0A139WFD4 A0A1S6Q5L3 F4X885 A0A158NBQ2 D6WT31 E2BCA4 A0A026WC60 A0A0L7QU49 A0A2J7Q9G1 A0A2P8Z4B7 A0A067RTL6 A0A0M9A518 E2A1P1 A0A087ZZN8 A0A195FD54 A0A151X9I5 A0A195BG81 A0A195DS04 A0A1Y1N7Z0 A0A0J7K3A1 K7IUK2 J3JTS6 N6TD48 A0A1B6CPF7 A0A232F5X5 A0A0P5IE39 A0A154P3U7 E9HGR7 A0A0P5G816 A0A0P4ZDQ8 A0A0P6E054 A0A0P5G0X2 A0A0N8C4N2 A0A0P6FYK7 A0A0P6EKE3 A0A0P5S5S9 A0A0P5JKP0 A0A0P5ASF6 A0A1B6EX92 A0A0P5HLG0 A0A0P6DJ38 A0A1B6H9C8 A0A0P6GW73 A0A0P5VCU5 A0A0P5KUL8 A0A0P5NHZ6 A0A0P4XSD7 A0A1B6L3Y8 T1JK43 A0A0P5SI01 A0A0P5G1X0 A0A0K8RHF0 A0A0P5V5N8 A0A0P5NQ08 T1DEV5 A0A0P5A6A2 Q1HQN1 A0A182GLM4 A0A023GGP0 G3MMH6 A0A1E1X441 E0VDI1 A0A2R5LG22 A0A0P6G830 A0A293MYB5 A0A023FK49 A0A023FXX5 A0A3S3PEI0 A0A1S4JRU4 A0A0P5SHE8 A0A310SRT7 A0A131XDD5 A0A226EMH8 A0A1D2MQC3 A0A224YRP6 A0A131Z3A6 L7M5R1 V4AEP3 B0WTL4 U5EMA6 A0A023EN66 S4REA7 A0A1Z5KXE3 A0A1Q3FDE8 C3ZPP8 A0A0P5VYK8 Q3TUI9 A0A1S3GET8
A0A2W1B5X0 A0A2A4JKH8 A0A194Q2V7 A0A212FER0 E9IFI5 A0A151IK79 A0A139WFD4 A0A1S6Q5L3 F4X885 A0A158NBQ2 D6WT31 E2BCA4 A0A026WC60 A0A0L7QU49 A0A2J7Q9G1 A0A2P8Z4B7 A0A067RTL6 A0A0M9A518 E2A1P1 A0A087ZZN8 A0A195FD54 A0A151X9I5 A0A195BG81 A0A195DS04 A0A1Y1N7Z0 A0A0J7K3A1 K7IUK2 J3JTS6 N6TD48 A0A1B6CPF7 A0A232F5X5 A0A0P5IE39 A0A154P3U7 E9HGR7 A0A0P5G816 A0A0P4ZDQ8 A0A0P6E054 A0A0P5G0X2 A0A0N8C4N2 A0A0P6FYK7 A0A0P6EKE3 A0A0P5S5S9 A0A0P5JKP0 A0A0P5ASF6 A0A1B6EX92 A0A0P5HLG0 A0A0P6DJ38 A0A1B6H9C8 A0A0P6GW73 A0A0P5VCU5 A0A0P5KUL8 A0A0P5NHZ6 A0A0P4XSD7 A0A1B6L3Y8 T1JK43 A0A0P5SI01 A0A0P5G1X0 A0A0K8RHF0 A0A0P5V5N8 A0A0P5NQ08 T1DEV5 A0A0P5A6A2 Q1HQN1 A0A182GLM4 A0A023GGP0 G3MMH6 A0A1E1X441 E0VDI1 A0A2R5LG22 A0A0P6G830 A0A293MYB5 A0A023FK49 A0A023FXX5 A0A3S3PEI0 A0A1S4JRU4 A0A0P5SHE8 A0A310SRT7 A0A131XDD5 A0A226EMH8 A0A1D2MQC3 A0A224YRP6 A0A131Z3A6 L7M5R1 V4AEP3 B0WTL4 U5EMA6 A0A023EN66 S4REA7 A0A1Z5KXE3 A0A1Q3FDE8 C3ZPP8 A0A0P5VYK8 Q3TUI9 A0A1S3GET8
PDB
6AVO
E-value=1.96325e-104,
Score=966
Ontologies
GO
Topology
Subcellular location
Nucleus
Cytoplasm
Cytoplasm
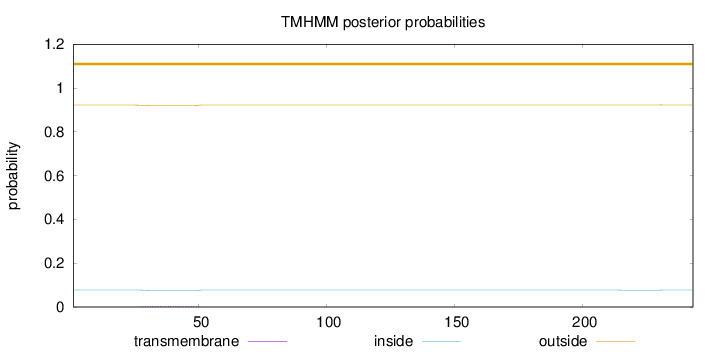
Length:
243
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0970099999999999
Exp number, first 60 AAs:
0.09541
Total prob of N-in:
0.07766
outside
1 - 243
Population Genetic Test Statistics
Pi
273.653854
Theta
223.273863
Tajima's D
0.705439
CLR
0
CSRT
0.581120943952802
Interpretation
Uncertain
