Gene
KWMTBOMO16257
Pre Gene Modal
BGIBMGA013994
Annotation
PREDICTED:_juvenile_hormone_epoxide_hydrolase-like_protein_3_isoform_X1_[Bombyx_mori]
Full name
Glutathione peroxidase
+ More
Epoxide hydrolase
Juvenile hormone epoxide hydrolase
Juvenile hormone epoxide hydrolase 1
Epoxide hydrolase
Juvenile hormone epoxide hydrolase
Juvenile hormone epoxide hydrolase 1
Alternative Name
CfEH1
Juvenile hormone epoxide hydrolase I
Juvenile hormone-specific epoxide hydrolase I
Juvenile hormone epoxide hydrolase I
Juvenile hormone-specific epoxide hydrolase I
Location in the cell
Cytoplasmic Reliability : 1.675 Mitochondrial Reliability : 1.039
Sequence
CDS
ATGAATTCCAACGAAAAAAATAAGTCATTCAAATCATTTATTCTCATCACTGTCATACAAGTGCTCAGCGCATATTATCTGTATCGCTTTTTCTTCGTACCACCTGATATGCCAGCACTTGACACTGAACAATGGTGGGGCCCGTACCCTATGGACTTGGACCACGATAAATCAATAAGACCGCTAACGATAGAATTTTCTGATGTGATGATAAACGATCTACGCGAGCGACTTCTCCACCGAAGACCCCTACAGCCACCCTTAGAAAATGTTGGATTCACATACGGCTTCAACCCAAGATATTTAAACCAAATAATGGAGTATTGGCATAACAAATACAACTTTAAAGAGAGGGAACAGTTTTTGAACCAATACGATCACTTTGTAACCAATATCCAGGGTTTGGACATACATTTCATGCATATAAAACCTAAGAACATTGCTGGTCTTGAGGTGGTTCCTCTAATCATGCTGCATGGATGGCCCGGTTCATTCAGGGAATTCTATGATGTCATCCCATATTTAATGGCAGAACAGCCAGAACAAAAAATCGCTTTTGAAATCATCGTGCCAAGCTTACCCGGTTTTGGTTACTCACAGGCCTCGGTGAGACCAGGTTTGGGTCCAGCTGAAATGGCGGTGATCATCAACAATCTCATGAAGAGGATCGGCCATGATAAATATTACATACAAGGTGGAGACCTAGGGCACACTGTTGGCTCCATCATTGCTACTGCATTCCCTGAAAACGTATTAGGATTTCACACAAATTTCCCAGTGCTTCTATCTCATAGAGCTGCTCTCTTATATATAACTTGTGGCTCTTTGATTCCATCTCTAATTGAAACGAAAGAATTGCAACCCCGCCTGTACCCGCTATCCGAGCACTGGTCTAGACTCATTGAGGAGTCAGGGTACATGCACATTCAAGCCACAAAACCCGAGACTGTCGGTGTGGGACTATCAGACTCGCCAGCAGGCCTAGCAGCCTACATTCTAGAAAAGTTTTCAACTTGGACAAATCCTGATAACAAGAAGGCCGGTGACGGAAACCTCTTGAAGAAGTTCTCTTTGAATCAGCTATTGGATAATCTGATGATATACTGGGCCACTAACACTGTCACAACCAGCATGAGGGTGTATGCTGAATCATTTAATGAAAAACAAAACTTTCTTATTTTCGACAGGATACCAACAAATGTTCCGACGTGGGGTATAAAATTCAAGTATGAACTGATGTACCAACCGGACACAGCATTAAGTTTCAAATACAAGAAGTACTTGCAGAGCACTGTGGTGGAGGACGGTGGTCACTTCGCTGCCATGGAATATCCTGAATTGATGGCCAAAGATATCATACAAGCTGTGATTAAATTTCAAAAATTTTGGAATTTCGAAAAACAAGAATCTAACGAAACGCCGCCTCTCCACGAGACCGCGAGAACAGTATACGAATTTACAGTTAAATCAATAAACGGACGCGACGTCAAGCTGTCAGATTACAAAGGGAACGTGCTGCTCATTGTCAATGTAGCCTCTCAGTGCGGCCTTACGGCCACCAACTATCAACAACTCAACGAGCTACACGAAAAGTACCACCAGAAAGGTCTACGTATACTGGCCTTCCCGTGCAACCAGTTCAACGGCCAAGAACCAGGTACAGCTAAGGACATTCTGAACTTCACAAAAGACAGAGGCGTTAAATTTGATCTGTTTGAAAAAGTCGATGTGAACGGTGACAACGCGCATCCGCTTTGGAAGTTTCTGAAGAAGGCGCAGAGCGGGACGATTGGTGATTTTATCAAGTGGAACTTTAGTAAGTTCGTAGTCGACAGAAACGGCGTGCCCGTCGAGAGATACGCGCCGCATGTCAATCCACTGGATCTGGAAAAGGATTTGGCCAAATATTGGTAA
Protein
MNSNEKNKSFKSFILITVIQVLSAYYLYRFFFVPPDMPALDTEQWWGPYPMDLDHDKSIRPLTIEFSDVMINDLRERLLHRRPLQPPLENVGFTYGFNPRYLNQIMEYWHNKYNFKEREQFLNQYDHFVTNIQGLDIHFMHIKPKNIAGLEVVPLIMLHGWPGSFREFYDVIPYLMAEQPEQKIAFEIIVPSLPGFGYSQASVRPGLGPAEMAVIINNLMKRIGHDKYYIQGGDLGHTVGSIIATAFPENVLGFHTNFPVLLSHRAALLYITCGSLIPSLIETKELQPRLYPLSEHWSRLIEESGYMHIQATKPETVGVGLSDSPAGLAAYILEKFSTWTNPDNKKAGDGNLLKKFSLNQLLDNLMIYWATNTVTTSMRVYAESFNEKQNFLIFDRIPTNVPTWGIKFKYELMYQPDTALSFKYKKYLQSTVVEDGGHFAAMEYPELMAKDIIQAVIKFQKFWNFEKQESNETPPLHETARTVYEFTVKSINGRDVKLSDYKGNVLLIVNVASQCGLTATNYQQLNELHEKYHQKGLRILAFPCNQFNGQEPGTAKDILNFTKDRGVKFDLFEKVDVNGDNAHPLWKFLKKAQSGTIGDFIKWNFSKFVVDRNGVPVERYAPHVNPLDLEKDLAKYW
Summary
Description
Catalyzes juvenile hormone hydrolysis.
Catalyzes juvenile hormone hydrolysis. Degrades juvenile hormone III (JH III) about 3 times and 5 times slower than juvenile hormone I (JH I) and II (JH II), respectively. Degrades cis-stilbene oxide and trans-stilbene oxide about 18 and 43 times slower than JH III, respectively.
Catalyzes juvenile hormone hydrolysis. Degrades juvenile hormone III (JH III) about 3 times and 5 times slower than juvenile hormone I (JH I) and II (JH II), respectively. Degrades cis-stilbene oxide and trans-stilbene oxide about 18 and 43 times slower than JH III, respectively.
Catalytic Activity
1-(4-methoxyphenyl)-N-methyl-N-[(3-methyloxetan-3-yl)methyl]methanamine + H2O = 2-{[(4-methoxybenzyl)(methyl)amino]methyl}-2-methylpropane-1,3-diol
cis-stilbene oxide + H2O = (1R,2R)-hydrobenzoin
cis-stilbene oxide + H2O = (1R,2R)-hydrobenzoin
Biophysicochemical Properties
0.52 uM for juvenile hormone III
Subunit
Homodimer.
Similarity
Belongs to the glutathione peroxidase family.
Belongs to the peptidase S33 family.
Belongs to the peptidase S33 family.
Keywords
3D-structure
Aromatic hydrocarbons catabolism
Complete proteome
Endoplasmic reticulum
Hydrolase
Membrane
Microsome
Reference proteome
Transmembrane
Transmembrane helix
Direct protein sequencing
Feature
chain Epoxide hydrolase
Uniprot
H9JWT0
C6KWM7
A0A0N0PD06
A0A2W1BCV7
A0A088M9L0
A0A2A4JKY8
+ More
A0A3S2NYE4 A0A194PYB3 A0A1V0M8A5 A0A2H1VS44 A0A0P0ELB6 H9JWL6 Q6U6J0 A0A2W1B6Y8 O44124 Q1W696 Q94806 A0A0N0PEI4 A0A212F9W9 A0A3S2LEJ8 A0A2A4JV24 V5G602 D6WGJ2 D0W012 Q8MZR6 D0W015 A0A3S2NQU9 A0A0L7LT60 A0A2C9DK11 A0A2H1VAR3 A0A1W5KJY8 J3JY41 E9HGT3 A0A182ME36 A0A3S2P9A9 A0A0T6BDK1 R4G4K1 A0A1I8JUR7 A0A0F6SIA8 A0A3L8DLY6 A0A0N0P9P2 F4WV40 A0A0P5ZLI5 A0A1Y1K079 A0A1Y1K345 T1PCN6 A0A023ESJ9 C6KWM8 A0A0P5IQR9 A0A023EUU0 A0A2M4BMN3 A0A2M4BM54
A0A3S2NYE4 A0A194PYB3 A0A1V0M8A5 A0A2H1VS44 A0A0P0ELB6 H9JWL6 Q6U6J0 A0A2W1B6Y8 O44124 Q1W696 Q94806 A0A0N0PEI4 A0A212F9W9 A0A3S2LEJ8 A0A2A4JV24 V5G602 D6WGJ2 D0W012 Q8MZR6 D0W015 A0A3S2NQU9 A0A0L7LT60 A0A2C9DK11 A0A2H1VAR3 A0A1W5KJY8 J3JY41 E9HGT3 A0A182ME36 A0A3S2P9A9 A0A0T6BDK1 R4G4K1 A0A1I8JUR7 A0A0F6SIA8 A0A3L8DLY6 A0A0N0P9P2 F4WV40 A0A0P5ZLI5 A0A1Y1K079 A0A1Y1K345 T1PCN6 A0A023ESJ9 C6KWM8 A0A0P5IQR9 A0A023EUU0 A0A2M4BMN3 A0A2M4BM54
EC Number
3.3.2.9
Pubmed
EMBL
BABH01018608
AB293556
BAH97090.1
KQ460417
KPJ14867.1
KZ150476
+ More
PZC70740.1 KJ579233 AIN34709.1 NWSH01001212 PCG72103.1 RSAL01000025 RVE52162.1 KQ459588 KPI97739.1 KU755500 ARD71210.1 ODYU01004100 SOQ43618.1 KT261710 ALJ30250.1 BABH01018379 AY377854 AB362775 AAQ87024.1 BAF81491.1 KZ150418 PZC70941.1 AF035482 AAB88192.1 DQ431847 ABD85119.1 U73680 AAB18243.1 KQ459764 KPJ20089.1 AGBW02009550 OWR50536.1 RSAL01000177 RVE45005.1 NWSH01000520 PCG75887.1 GALX01002976 JAB65490.1 KQ971321 EFA00568.1 AB534186 BAI49687.1 AF503908 AAM22694.1 AB534189 BAI49690.1 RSAL01000008 RVE53920.1 JTDY01000179 KOB78426.1 KX855995 ARE68677.1 ODYU01001536 SOQ37891.1 KU941586 AMR44689.1 BT128165 AEE63126.1 GL732643 EFX69015.1 AXCM01002766 RVE45007.1 LJIG01001532 KRT85408.1 ACPB03008739 GAHY01001290 JAA76220.1 KP271046 AKF11871.1 QOIP01000006 RLU21450.1 KQ459547 KPJ00030.1 GL888384 EGI61928.1 GDIP01250763 GDIP01043274 JAM60441.1 GEZM01096421 JAV54922.1 GEZM01096422 JAV54921.1 KA645900 AFP60529.1 GAPW01001360 JAC12238.1 AB293557 BAH97091.1 GDIQ01210360 GDIQ01190372 JAK41365.1 GAPW01001359 JAC12239.1 GGFJ01005082 MBW54223.1 GGFJ01005028 MBW54169.1
PZC70740.1 KJ579233 AIN34709.1 NWSH01001212 PCG72103.1 RSAL01000025 RVE52162.1 KQ459588 KPI97739.1 KU755500 ARD71210.1 ODYU01004100 SOQ43618.1 KT261710 ALJ30250.1 BABH01018379 AY377854 AB362775 AAQ87024.1 BAF81491.1 KZ150418 PZC70941.1 AF035482 AAB88192.1 DQ431847 ABD85119.1 U73680 AAB18243.1 KQ459764 KPJ20089.1 AGBW02009550 OWR50536.1 RSAL01000177 RVE45005.1 NWSH01000520 PCG75887.1 GALX01002976 JAB65490.1 KQ971321 EFA00568.1 AB534186 BAI49687.1 AF503908 AAM22694.1 AB534189 BAI49690.1 RSAL01000008 RVE53920.1 JTDY01000179 KOB78426.1 KX855995 ARE68677.1 ODYU01001536 SOQ37891.1 KU941586 AMR44689.1 BT128165 AEE63126.1 GL732643 EFX69015.1 AXCM01002766 RVE45007.1 LJIG01001532 KRT85408.1 ACPB03008739 GAHY01001290 JAA76220.1 KP271046 AKF11871.1 QOIP01000006 RLU21450.1 KQ459547 KPJ00030.1 GL888384 EGI61928.1 GDIP01250763 GDIP01043274 JAM60441.1 GEZM01096421 JAV54922.1 GEZM01096422 JAV54921.1 KA645900 AFP60529.1 GAPW01001360 JAC12238.1 AB293557 BAH97091.1 GDIQ01210360 GDIQ01190372 JAK41365.1 GAPW01001359 JAC12239.1 GGFJ01005082 MBW54223.1 GGFJ01005028 MBW54169.1
Proteomes
Interpro
IPR029759
GPX_AS
+ More
IPR029760 GPX_CS
IPR000889 Glutathione_peroxidase
IPR029058 AB_hydrolase
IPR036249 Thioredoxin-like_sf
IPR010497 Epoxide_hydro_N
IPR016292 Epoxide_hydrolase
IPR000639 Epox_hydrolase-like
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR029760 GPX_CS
IPR000889 Glutathione_peroxidase
IPR029058 AB_hydrolase
IPR036249 Thioredoxin-like_sf
IPR010497 Epoxide_hydro_N
IPR016292 Epoxide_hydrolase
IPR000639 Epox_hydrolase-like
IPR005135 Endo/exonuclease/phosphatase
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
Gene 3D
ProteinModelPortal
H9JWT0
C6KWM7
A0A0N0PD06
A0A2W1BCV7
A0A088M9L0
A0A2A4JKY8
+ More
A0A3S2NYE4 A0A194PYB3 A0A1V0M8A5 A0A2H1VS44 A0A0P0ELB6 H9JWL6 Q6U6J0 A0A2W1B6Y8 O44124 Q1W696 Q94806 A0A0N0PEI4 A0A212F9W9 A0A3S2LEJ8 A0A2A4JV24 V5G602 D6WGJ2 D0W012 Q8MZR6 D0W015 A0A3S2NQU9 A0A0L7LT60 A0A2C9DK11 A0A2H1VAR3 A0A1W5KJY8 J3JY41 E9HGT3 A0A182ME36 A0A3S2P9A9 A0A0T6BDK1 R4G4K1 A0A1I8JUR7 A0A0F6SIA8 A0A3L8DLY6 A0A0N0P9P2 F4WV40 A0A0P5ZLI5 A0A1Y1K079 A0A1Y1K345 T1PCN6 A0A023ESJ9 C6KWM8 A0A0P5IQR9 A0A023EUU0 A0A2M4BMN3 A0A2M4BM54
A0A3S2NYE4 A0A194PYB3 A0A1V0M8A5 A0A2H1VS44 A0A0P0ELB6 H9JWL6 Q6U6J0 A0A2W1B6Y8 O44124 Q1W696 Q94806 A0A0N0PEI4 A0A212F9W9 A0A3S2LEJ8 A0A2A4JV24 V5G602 D6WGJ2 D0W012 Q8MZR6 D0W015 A0A3S2NQU9 A0A0L7LT60 A0A2C9DK11 A0A2H1VAR3 A0A1W5KJY8 J3JY41 E9HGT3 A0A182ME36 A0A3S2P9A9 A0A0T6BDK1 R4G4K1 A0A1I8JUR7 A0A0F6SIA8 A0A3L8DLY6 A0A0N0P9P2 F4WV40 A0A0P5ZLI5 A0A1Y1K079 A0A1Y1K345 T1PCN6 A0A023ESJ9 C6KWM8 A0A0P5IQR9 A0A023EUU0 A0A2M4BMN3 A0A2M4BM54
PDB
4QLA
E-value=1.64658e-137,
Score=1256
Ontologies
GO
Topology
Subcellular location
Endoplasmic reticulum membrane
Microsome membrane
Microsome membrane
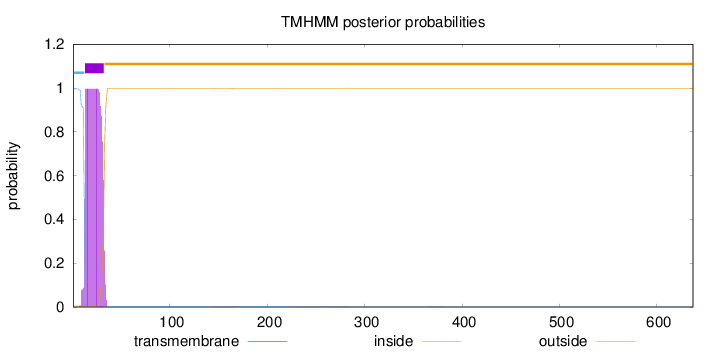
Length:
637
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.25292
Exp number, first 60 AAs:
20.1975
Total prob of N-in:
0.99548
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 32
outside
33 - 637
Population Genetic Test Statistics
Pi
259.077384
Theta
168.849024
Tajima's D
1.604172
CLR
0.026608
CSRT
0.810559472026399
Interpretation
Uncertain
