Gene
KWMTBOMO16255
Pre Gene Modal
BGIBMGA013993
Annotation
PREDICTED:_uncharacterized_protein_LOC105841315_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.564 Nuclear Reliability : 1.705
Sequence
CDS
ATGAACCTGCGTTCTAAGGTTTGGGCTGCCAAGACTTCGCTCAAGGGCTCAGGAATAACACTCTCCGAGTTCTTGACTAAAAGACGTCATGATATTTTTATGTCTGCACGTCAACGCTTCGGAGTAACCAGGTGCTGGACTCGGGACGGCTGCGTTTACGTTCAGGGATCGGATGGGGTGCGACATCGTGTCGAAGTGATGTCTGATCTGGATGCTCTTGCCAAGACGACTATGGCGGAAGATGCTGCCCAAAAGAGGGCACAAACCGGCAAGGACACAAAGGAGTCTGCAGCTGCGACCAGACCAAAACGTGCGGCTGCGAACAAGAAGTAG
Protein
MNLRSKVWAAKTSLKGSGITLSEFLTKRRHDIFMSARQRFGVTRCWTRDGCVYVQGSDGVRHRVEVMSDLDALAKTTMAEDAAQKRAQTGKDTKESAAATRPKRAAANKK
Summary
Uniprot
H9JWS9
H9JND1
A0A2A4ITX7
A0A2A4J0P1
A0A0L7LEM3
A0A2A4JPZ6
+ More
A0A2A4JP60 A0A0L7K3B3 A0A0L7KNA4 A0A2W1C1G2 A0A2H1W7N3 A0A2H1X0E0 A0A0L7K1Y0 A0A0L7L7Q1 A0A0L7KMJ9 A0A0L7K490 A0A0L7KIU7 A0A0L7L318 A0A0L7KW56 A0A2W1BKL8 A0A2A4JRK8 A0A2A4JAE4 A0A0L7KWV6 A0A2W1BHY6 A0A2A4JIA9 A0A2A4ISJ1 A0A2A4JVB8 A0A0L7KQ82 A0A2W1B638 A0A0L7LT08 A0A0L7KL53 A0A3Q0J8Z8 A0A3Q0IRU1 A0A0L7LN09
A0A2A4JP60 A0A0L7K3B3 A0A0L7KNA4 A0A2W1C1G2 A0A2H1W7N3 A0A2H1X0E0 A0A0L7K1Y0 A0A0L7L7Q1 A0A0L7KMJ9 A0A0L7K490 A0A0L7KIU7 A0A0L7L318 A0A0L7KW56 A0A2W1BKL8 A0A2A4JRK8 A0A2A4JAE4 A0A0L7KWV6 A0A2W1BHY6 A0A2A4JIA9 A0A2A4ISJ1 A0A2A4JVB8 A0A0L7KQ82 A0A2W1B638 A0A0L7LT08 A0A0L7KL53 A0A3Q0J8Z8 A0A3Q0IRU1 A0A0L7LN09
EMBL
BABH01018592
BABH01019446
BABH01019447
BABH01019448
BABH01019449
BABH01019450
+ More
NWSH01007488 PCG62946.1 NWSH01004248 PCG65306.1 JTDY01001444 KOB73845.1 NWSH01000800 PCG74117.1 NWSH01000964 PCG73374.1 JTDY01012534 KOB52261.1 JTDY01008093 KOB64747.1 KZ149916 PZC77943.1 ODYU01006869 SOQ49099.1 ODYU01012456 SOQ58773.1 JTDY01016995 KOB51870.1 JTDY01002460 KOB71369.1 JTDY01009052 KOB64154.1 JTDY01010801 JTDY01009435 KOB57999.1 KOB63512.1 JTDY01009601 KOB63242.1 JTDY01003374 KOB69654.1 JTDY01005020 KOB67468.1 KZ150083 PZC73827.1 NWSH01000709 PCG74665.1 NWSH01002122 PCG69097.1 JTDY01004815 JTDY01001265 KOB67717.1 KOB74342.1 KZ150113 PZC73314.1 NWSH01001465 PCG71162.1 PCG71163.1 NWSH01008089 PCG62701.1 NWSH01000574 PCG75564.1 JTDY01007409 KOB65221.1 KZ150731 PZC70355.1 JTDY01000147 KOB78598.1 JTDY01009138 KOB64057.1 JTDY01000521 KOB76815.1
NWSH01007488 PCG62946.1 NWSH01004248 PCG65306.1 JTDY01001444 KOB73845.1 NWSH01000800 PCG74117.1 NWSH01000964 PCG73374.1 JTDY01012534 KOB52261.1 JTDY01008093 KOB64747.1 KZ149916 PZC77943.1 ODYU01006869 SOQ49099.1 ODYU01012456 SOQ58773.1 JTDY01016995 KOB51870.1 JTDY01002460 KOB71369.1 JTDY01009052 KOB64154.1 JTDY01010801 JTDY01009435 KOB57999.1 KOB63512.1 JTDY01009601 KOB63242.1 JTDY01003374 KOB69654.1 JTDY01005020 KOB67468.1 KZ150083 PZC73827.1 NWSH01000709 PCG74665.1 NWSH01002122 PCG69097.1 JTDY01004815 JTDY01001265 KOB67717.1 KOB74342.1 KZ150113 PZC73314.1 NWSH01001465 PCG71162.1 PCG71163.1 NWSH01008089 PCG62701.1 NWSH01000574 PCG75564.1 JTDY01007409 KOB65221.1 KZ150731 PZC70355.1 JTDY01000147 KOB78598.1 JTDY01009138 KOB64057.1 JTDY01000521 KOB76815.1
Proteomes
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JWS9
H9JND1
A0A2A4ITX7
A0A2A4J0P1
A0A0L7LEM3
A0A2A4JPZ6
+ More
A0A2A4JP60 A0A0L7K3B3 A0A0L7KNA4 A0A2W1C1G2 A0A2H1W7N3 A0A2H1X0E0 A0A0L7K1Y0 A0A0L7L7Q1 A0A0L7KMJ9 A0A0L7K490 A0A0L7KIU7 A0A0L7L318 A0A0L7KW56 A0A2W1BKL8 A0A2A4JRK8 A0A2A4JAE4 A0A0L7KWV6 A0A2W1BHY6 A0A2A4JIA9 A0A2A4ISJ1 A0A2A4JVB8 A0A0L7KQ82 A0A2W1B638 A0A0L7LT08 A0A0L7KL53 A0A3Q0J8Z8 A0A3Q0IRU1 A0A0L7LN09
A0A2A4JP60 A0A0L7K3B3 A0A0L7KNA4 A0A2W1C1G2 A0A2H1W7N3 A0A2H1X0E0 A0A0L7K1Y0 A0A0L7L7Q1 A0A0L7KMJ9 A0A0L7K490 A0A0L7KIU7 A0A0L7L318 A0A0L7KW56 A0A2W1BKL8 A0A2A4JRK8 A0A2A4JAE4 A0A0L7KWV6 A0A2W1BHY6 A0A2A4JIA9 A0A2A4ISJ1 A0A2A4JVB8 A0A0L7KQ82 A0A2W1B638 A0A0L7LT08 A0A0L7KL53 A0A3Q0J8Z8 A0A3Q0IRU1 A0A0L7LN09
Ontologies
PANTHER
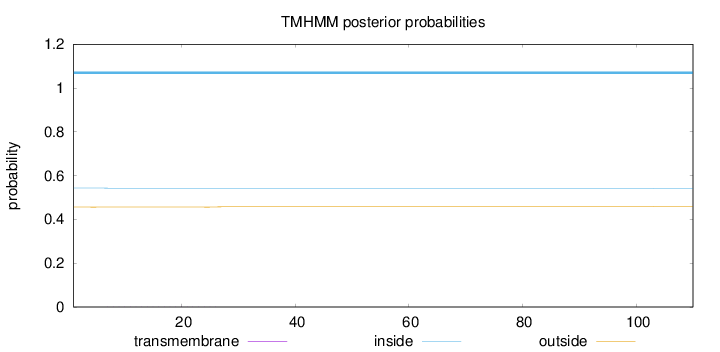
Topology
Length:
110
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02366
Exp number, first 60 AAs:
0.02366
Total prob of N-in:
0.54362
inside
1 - 110
Population Genetic Test Statistics
Pi
244.108683
Theta
129.76716
Tajima's D
2.645939
CLR
0
CSRT
0.960501974901255
Interpretation
Uncertain
