Pre Gene Modal
BGIBMGA013902
Annotation
PREDICTED:_adenine_phosphoribosyltransferase_isoform_X3_[Bombyx_mori]
Full name
Adenine phosphoribosyltransferase
Location in the cell
Cytoplasmic Reliability : 2.435
Sequence
CDS
ATGAATAAAGACTACGAAAATAAAGTAGTCGTTCTAAAGAGTAAGATAAAAAGCTTTCCCGATTTTCCAAAAAAGGGAATCTTATTCTGGGACATATTTTCGGCAATATCAGATGGCAACATATGCAAACTATTGCAATCACTATTAGTTGATTGTATACGTACAAAATTTCCCGACGTAGAAGCTGTAGTTGGACTAGAATCTCGAGGATTCCTGTTCTCTTTCTCGCTTGCTTCAGAATTAGGAATCAGCTGTCTTTCAGTCAGGAAGAAGGGAAAGCTGCCAGGAGATGTAATATCTTATGAATATGATTTAGAATATGGTTCGGACACATTGGAAATCCAAAAAGATAAAATACGGCCAGGTTTAAAATGTCTGCTAGTTGATGATTTAATAGCAACTGGCGGTTCCATAACGGCCGCGACAAAATTACTGCAAACATGCGGTGCCGAAGTTCTTGGCTGTTTGGTCATCATTGAACTTAAAAGTTTGAAAGGAAGAGAGAACATTCCTGAAGGTATACCAGTGCATTCCTTGATTCAATATGAATAG
Protein
MNKDYENKVVVLKSKIKSFPDFPKKGILFWDIFSAISDGNICKLLQSLLVDCIRTKFPDVEAVVGLESRGFLFSFSLASELGISCLSVRKKGKLPGDVISYEYDLEYGSDTLEIQKDKIRPGLKCLLVDDLIATGGSITAATKLLQTCGAEVLGCLVIIELKSLKGRENIPEGIPVHSLIQYE
Summary
Description
Catalyzes a salvage reaction resulting in the formation of AMP, that is energically less costly than de novo synthesis.
Catalytic Activity
AMP + diphosphate = 5-phospho-alpha-D-ribose 1-diphosphate + adenine
Subunit
Homodimer.
Similarity
Belongs to the purine/pyrimidine phosphoribosyltransferase family.
Keywords
Complete proteome
Cytoplasm
Glycosyltransferase
Purine salvage
Reference proteome
Transferase
Feature
chain Adenine phosphoribosyltransferase
Uniprot
H9JWI8
A0A2W1BR32
A0A0N0P9T0
A0A0N1IQ42
A0A3S2TPQ1
A0A212F5X8
+ More
A0A2H1W4Z6 A0A1B0AVU9 A0A1A9XRA0 D3TRH0 A0A1B0A291 A0A1B0FDG9 B4MXH1 A0A1A9VFE5 A0A1A9WGQ3 A0A0L0CKH2 Q1HQP5 B4KWW4 W8BVB6 A0A023EI33 A0A182GDY2 A0A0K8TSL2 A0A0M4ECZ8 A0A336LYK4 A0A034VRH8 A0A0K8WE68 A0A1Q3F7Q6 A0A0A1WX85 A0A1I8PVM9 A0A1L8DXR5 T1P9T4 A0A1B0CY73 B4PE88 B3NEX4 B4LH08 B4HW24 B0W403 B4IWQ2 B4QN03 A0A1W4W4M8 B4GS35 A0A182PMK4 X2J8L3 P12426 A0A182TII2 A0A182L305 Q7Q6Q8 A0A182HWL7 A0A2M4CLT0 A0A2M4CMW2 W5JSA6 A0A2M4C0G3 A0A182XMQ9 A0A182YD60 O44095 A0A084WDM3 A0A3B0JRX2 P54363 A0A182K8C4 A0A2M4C0H2 A0A182NIC5 A0A182MVT3 A0A2M4C0H3 A0A182WKV2 A0A2M3ZIG3 A0A182QN99 A0A2M4AN20 A0A2M3ZA44 A0A182T3T6 U5EJG9 A0A226F3J2 A0A0T6B7M4 B3M785 B5FX92 A0A0Q9XMW8 A0A1Y1MCW1 A0A2R7X0S6 E9IH41 A0A1J1J515 A0A026X030 A0A3S3PL76 A0A091HKD1 A0A1B0CP83 W5KUD4 A0A195C7F2 A0A158NTX8 D6WSW8 A0A182FE58 A0A2P8XH02 A0A0D2WKN8 A0A0B7AU19 F4WUB2 A0A3Q0SKE6 A0A3B4FJ72 A0A3P8NYD3 A0A3P9CR19 A0A0F8B451 I3K5Y7
A0A2H1W4Z6 A0A1B0AVU9 A0A1A9XRA0 D3TRH0 A0A1B0A291 A0A1B0FDG9 B4MXH1 A0A1A9VFE5 A0A1A9WGQ3 A0A0L0CKH2 Q1HQP5 B4KWW4 W8BVB6 A0A023EI33 A0A182GDY2 A0A0K8TSL2 A0A0M4ECZ8 A0A336LYK4 A0A034VRH8 A0A0K8WE68 A0A1Q3F7Q6 A0A0A1WX85 A0A1I8PVM9 A0A1L8DXR5 T1P9T4 A0A1B0CY73 B4PE88 B3NEX4 B4LH08 B4HW24 B0W403 B4IWQ2 B4QN03 A0A1W4W4M8 B4GS35 A0A182PMK4 X2J8L3 P12426 A0A182TII2 A0A182L305 Q7Q6Q8 A0A182HWL7 A0A2M4CLT0 A0A2M4CMW2 W5JSA6 A0A2M4C0G3 A0A182XMQ9 A0A182YD60 O44095 A0A084WDM3 A0A3B0JRX2 P54363 A0A182K8C4 A0A2M4C0H2 A0A182NIC5 A0A182MVT3 A0A2M4C0H3 A0A182WKV2 A0A2M3ZIG3 A0A182QN99 A0A2M4AN20 A0A2M3ZA44 A0A182T3T6 U5EJG9 A0A226F3J2 A0A0T6B7M4 B3M785 B5FX92 A0A0Q9XMW8 A0A1Y1MCW1 A0A2R7X0S6 E9IH41 A0A1J1J515 A0A026X030 A0A3S3PL76 A0A091HKD1 A0A1B0CP83 W5KUD4 A0A195C7F2 A0A158NTX8 D6WSW8 A0A182FE58 A0A2P8XH02 A0A0D2WKN8 A0A0B7AU19 F4WUB2 A0A3Q0SKE6 A0A3B4FJ72 A0A3P8NYD3 A0A3P9CR19 A0A0F8B451 I3K5Y7
EC Number
2.4.2.7
Pubmed
19121390
28756777
26354079
22118469
20353571
17994087
+ More
26108605 17204158 17510324 24495485 24945155 26483478 26369729 25348373 25830018 25315136 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 3125085 8492806 12537569 20966253 12364791 14747013 17210077 20920257 23761445 25244985 9720289 24438588 15632085 17018643 28004739 21282665 24508170 30249741 25329095 21347285 18362917 19820115 29403074 21719571 25186727 25835551
26108605 17204158 17510324 24495485 24945155 26483478 26369729 25348373 25830018 25315136 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 3125085 8492806 12537569 20966253 12364791 14747013 17210077 20920257 23761445 25244985 9720289 24438588 15632085 17018643 28004739 21282665 24508170 30249741 25329095 21347285 18362917 19820115 29403074 21719571 25186727 25835551
EMBL
BABH01018578
KZ149970
PZC76075.1
KQ459166
KPJ03555.1
KQ459992
+ More
KPJ18602.1 RSAL01000029 RVE51766.1 AGBW02010113 OWR49142.1 ODYU01006364 SOQ48151.1 JXJN01004398 EZ424022 ADD20298.1 CCAG010014128 CH963876 EDW76740.1 JRES01000281 KNC32727.1 DQ440399 CH477368 ABF18432.1 EAT42485.1 CH933809 EDW18585.1 GAMC01001281 GAMC01001280 JAC05276.1 JXUM01011310 GAPW01004851 KQ560328 JAC08747.1 KXJ83016.1 JXUM01010024 KQ560297 KXJ83185.1 GDAI01000241 JAI17362.1 CP012525 ALC43384.1 UFQT01000305 SSX23062.1 GAKP01013878 JAC45074.1 GDHF01003164 JAI49150.1 GFDL01011468 JAV23577.1 GBXI01011157 JAD03135.1 GFDF01002834 JAV11250.1 KA645477 AFP60106.1 AJWK01035517 CM000159 EDW92926.1 CH954178 EDV50247.1 CH940647 EDW69498.1 CH480817 EDW50139.1 DS231833 EDS32300.1 CH916366 EDV96278.1 CM000363 CM002912 EDX08885.1 KMY96983.1 CH479188 EDW40570.1 AE014296 AHN57933.1 AHN57934.1 M18432 L06280 AY119066 AAAB01008960 EAA10726.4 APCN01001503 GGFL01002061 MBW66239.1 GGFL01002060 MBW66238.1 ADMH02000499 ETN66188.1 GGFJ01009676 MBW58817.1 AF025800 AAB87885.1 ATLV01023045 KE525339 KFB48317.1 OUUW01000009 SPP84884.1 L06281 CH379069 GGFJ01009672 MBW58813.1 AXCM01005962 GGFJ01009675 MBW58816.1 GGFM01007548 MBW28299.1 AXCN02000880 GGFK01008862 MBW42183.1 GGFM01004557 MBW25308.1 GANO01002238 JAB57633.1 LNIX01000001 OXA63486.1 LJIG01009329 KRT83297.1 CH902618 EDV38746.1 DQ213177 ACH43653.1 KRG06270.1 GEZM01038245 JAV81696.1 KK856295 PTY25407.1 GL763174 EFZ20096.1 CVRI01000067 CRL06900.1 KK107054 QOIP01000004 EZA61607.1 RLU23916.1 NCKU01006308 NCKU01006051 NCKU01005112 RWS03639.1 RWS03883.1 RWS05000.1 KL217481 KFO95649.1 AJWK01021534 KQ978143 KYM96804.1 ADTU01026194 KQ971352 EFA06669.1 PYGN01002161 PSN31287.1 AVFK01000008 KJB95179.1 HACG01036606 CEK83471.1 GL888355 EGI62220.1 KQ041719 KKF23533.1 AERX01042073 AERX01042074
KPJ18602.1 RSAL01000029 RVE51766.1 AGBW02010113 OWR49142.1 ODYU01006364 SOQ48151.1 JXJN01004398 EZ424022 ADD20298.1 CCAG010014128 CH963876 EDW76740.1 JRES01000281 KNC32727.1 DQ440399 CH477368 ABF18432.1 EAT42485.1 CH933809 EDW18585.1 GAMC01001281 GAMC01001280 JAC05276.1 JXUM01011310 GAPW01004851 KQ560328 JAC08747.1 KXJ83016.1 JXUM01010024 KQ560297 KXJ83185.1 GDAI01000241 JAI17362.1 CP012525 ALC43384.1 UFQT01000305 SSX23062.1 GAKP01013878 JAC45074.1 GDHF01003164 JAI49150.1 GFDL01011468 JAV23577.1 GBXI01011157 JAD03135.1 GFDF01002834 JAV11250.1 KA645477 AFP60106.1 AJWK01035517 CM000159 EDW92926.1 CH954178 EDV50247.1 CH940647 EDW69498.1 CH480817 EDW50139.1 DS231833 EDS32300.1 CH916366 EDV96278.1 CM000363 CM002912 EDX08885.1 KMY96983.1 CH479188 EDW40570.1 AE014296 AHN57933.1 AHN57934.1 M18432 L06280 AY119066 AAAB01008960 EAA10726.4 APCN01001503 GGFL01002061 MBW66239.1 GGFL01002060 MBW66238.1 ADMH02000499 ETN66188.1 GGFJ01009676 MBW58817.1 AF025800 AAB87885.1 ATLV01023045 KE525339 KFB48317.1 OUUW01000009 SPP84884.1 L06281 CH379069 GGFJ01009672 MBW58813.1 AXCM01005962 GGFJ01009675 MBW58816.1 GGFM01007548 MBW28299.1 AXCN02000880 GGFK01008862 MBW42183.1 GGFM01004557 MBW25308.1 GANO01002238 JAB57633.1 LNIX01000001 OXA63486.1 LJIG01009329 KRT83297.1 CH902618 EDV38746.1 DQ213177 ACH43653.1 KRG06270.1 GEZM01038245 JAV81696.1 KK856295 PTY25407.1 GL763174 EFZ20096.1 CVRI01000067 CRL06900.1 KK107054 QOIP01000004 EZA61607.1 RLU23916.1 NCKU01006308 NCKU01006051 NCKU01005112 RWS03639.1 RWS03883.1 RWS05000.1 KL217481 KFO95649.1 AJWK01021534 KQ978143 KYM96804.1 ADTU01026194 KQ971352 EFA06669.1 PYGN01002161 PSN31287.1 AVFK01000008 KJB95179.1 HACG01036606 CEK83471.1 GL888355 EGI62220.1 KQ041719 KKF23533.1 AERX01042073 AERX01042074
Proteomes
UP000005204
UP000053268
UP000053240
UP000283053
UP000007151
UP000092460
+ More
UP000092443 UP000092445 UP000092444 UP000007798 UP000078200 UP000091820 UP000037069 UP000008820 UP000009192 UP000069940 UP000249989 UP000092553 UP000095300 UP000095301 UP000092461 UP000002282 UP000008711 UP000008792 UP000001292 UP000002320 UP000001070 UP000000304 UP000192221 UP000008744 UP000075885 UP000000803 UP000075902 UP000075882 UP000007062 UP000075840 UP000000673 UP000076407 UP000076408 UP000030765 UP000268350 UP000001819 UP000075881 UP000075884 UP000075883 UP000075920 UP000075886 UP000075901 UP000198287 UP000007801 UP000183832 UP000053097 UP000279307 UP000285301 UP000054308 UP000018467 UP000078542 UP000005205 UP000007266 UP000069272 UP000245037 UP000032292 UP000007755 UP000261340 UP000261460 UP000265100 UP000265160 UP000005207
UP000092443 UP000092445 UP000092444 UP000007798 UP000078200 UP000091820 UP000037069 UP000008820 UP000009192 UP000069940 UP000249989 UP000092553 UP000095300 UP000095301 UP000092461 UP000002282 UP000008711 UP000008792 UP000001292 UP000002320 UP000001070 UP000000304 UP000192221 UP000008744 UP000075885 UP000000803 UP000075902 UP000075882 UP000007062 UP000075840 UP000000673 UP000076407 UP000076408 UP000030765 UP000268350 UP000001819 UP000075881 UP000075884 UP000075883 UP000075920 UP000075886 UP000075901 UP000198287 UP000007801 UP000183832 UP000053097 UP000279307 UP000285301 UP000054308 UP000018467 UP000078542 UP000005205 UP000007266 UP000069272 UP000245037 UP000032292 UP000007755 UP000261340 UP000261460 UP000265100 UP000265160 UP000005207
Interpro
SUPFAM
SSF53271
SSF53271
ProteinModelPortal
H9JWI8
A0A2W1BR32
A0A0N0P9T0
A0A0N1IQ42
A0A3S2TPQ1
A0A212F5X8
+ More
A0A2H1W4Z6 A0A1B0AVU9 A0A1A9XRA0 D3TRH0 A0A1B0A291 A0A1B0FDG9 B4MXH1 A0A1A9VFE5 A0A1A9WGQ3 A0A0L0CKH2 Q1HQP5 B4KWW4 W8BVB6 A0A023EI33 A0A182GDY2 A0A0K8TSL2 A0A0M4ECZ8 A0A336LYK4 A0A034VRH8 A0A0K8WE68 A0A1Q3F7Q6 A0A0A1WX85 A0A1I8PVM9 A0A1L8DXR5 T1P9T4 A0A1B0CY73 B4PE88 B3NEX4 B4LH08 B4HW24 B0W403 B4IWQ2 B4QN03 A0A1W4W4M8 B4GS35 A0A182PMK4 X2J8L3 P12426 A0A182TII2 A0A182L305 Q7Q6Q8 A0A182HWL7 A0A2M4CLT0 A0A2M4CMW2 W5JSA6 A0A2M4C0G3 A0A182XMQ9 A0A182YD60 O44095 A0A084WDM3 A0A3B0JRX2 P54363 A0A182K8C4 A0A2M4C0H2 A0A182NIC5 A0A182MVT3 A0A2M4C0H3 A0A182WKV2 A0A2M3ZIG3 A0A182QN99 A0A2M4AN20 A0A2M3ZA44 A0A182T3T6 U5EJG9 A0A226F3J2 A0A0T6B7M4 B3M785 B5FX92 A0A0Q9XMW8 A0A1Y1MCW1 A0A2R7X0S6 E9IH41 A0A1J1J515 A0A026X030 A0A3S3PL76 A0A091HKD1 A0A1B0CP83 W5KUD4 A0A195C7F2 A0A158NTX8 D6WSW8 A0A182FE58 A0A2P8XH02 A0A0D2WKN8 A0A0B7AU19 F4WUB2 A0A3Q0SKE6 A0A3B4FJ72 A0A3P8NYD3 A0A3P9CR19 A0A0F8B451 I3K5Y7
A0A2H1W4Z6 A0A1B0AVU9 A0A1A9XRA0 D3TRH0 A0A1B0A291 A0A1B0FDG9 B4MXH1 A0A1A9VFE5 A0A1A9WGQ3 A0A0L0CKH2 Q1HQP5 B4KWW4 W8BVB6 A0A023EI33 A0A182GDY2 A0A0K8TSL2 A0A0M4ECZ8 A0A336LYK4 A0A034VRH8 A0A0K8WE68 A0A1Q3F7Q6 A0A0A1WX85 A0A1I8PVM9 A0A1L8DXR5 T1P9T4 A0A1B0CY73 B4PE88 B3NEX4 B4LH08 B4HW24 B0W403 B4IWQ2 B4QN03 A0A1W4W4M8 B4GS35 A0A182PMK4 X2J8L3 P12426 A0A182TII2 A0A182L305 Q7Q6Q8 A0A182HWL7 A0A2M4CLT0 A0A2M4CMW2 W5JSA6 A0A2M4C0G3 A0A182XMQ9 A0A182YD60 O44095 A0A084WDM3 A0A3B0JRX2 P54363 A0A182K8C4 A0A2M4C0H2 A0A182NIC5 A0A182MVT3 A0A2M4C0H3 A0A182WKV2 A0A2M3ZIG3 A0A182QN99 A0A2M4AN20 A0A2M3ZA44 A0A182T3T6 U5EJG9 A0A226F3J2 A0A0T6B7M4 B3M785 B5FX92 A0A0Q9XMW8 A0A1Y1MCW1 A0A2R7X0S6 E9IH41 A0A1J1J515 A0A026X030 A0A3S3PL76 A0A091HKD1 A0A1B0CP83 W5KUD4 A0A195C7F2 A0A158NTX8 D6WSW8 A0A182FE58 A0A2P8XH02 A0A0D2WKN8 A0A0B7AU19 F4WUB2 A0A3Q0SKE6 A0A3B4FJ72 A0A3P8NYD3 A0A3P9CR19 A0A0F8B451 I3K5Y7
PDB
1ZN9
E-value=4.12108e-38,
Score=392
Ontologies
PATHWAY
GO
PANTHER
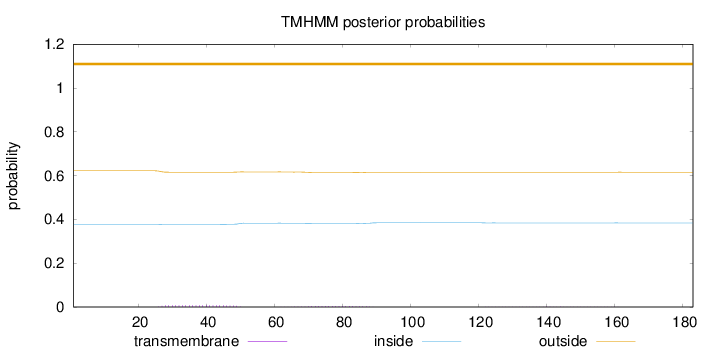
Topology
Subcellular location
Cytoplasm
Length:
183
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.27732
Exp number, first 60 AAs:
0.16697
Total prob of N-in:
0.37826
outside
1 - 183
Population Genetic Test Statistics
Pi
260.616602
Theta
195.564601
Tajima's D
1.091712
CLR
0.032155
CSRT
0.687515624218789
Interpretation
Uncertain
