Gene
KWMTBOMO16241
Pre Gene Modal
BGIBMGA013988
Annotation
PREDICTED:_alpha-N-acetylgalactosaminidase-like_[Papilio_xuthus]
Full name
Alpha-galactosidase
Location in the cell
Lysosomal Reliability : 2.252
Sequence
CDS
ATGGGGTGGATGTCGTGGGGGTATTACATGTGCGGCGCGGATTGCGAACAAAATCCGGACAAATGCTTAGATGAGAAATTAATTTTGTCAGTGGCAGACACATTTTACAATGAAGGTTACCAGGAAGCAGGTTTTGAATATATCATTATCGATGACTGCTGGTCCGAACATCACAGAGACAGAGACGGAAGACTTGTGCCTGATAGTCTGAGATTTCCTAATGGCATGAAGTACATTTCTGACTATATACATTCGCGTAATCTCAAATTCGGTATGTACACAAATGTAGCTGATGTCACTTGTATGAGATATCAAGGCTCAAAAGGACATTTTAAAGTCGACGCTGACACTTTTGCTCAATGGGGTGTCGATTATATCAAAGTGGATGGGTGTTTTGTGGGAGAACAGTACTTAAATAATGCATACATACATTTTGGTGGGTATTTAAACAAAACCAATAGACCTATGGTATATTCATGTAGCTGGCCCTATTACATAGAGTTTATACACAATAAGAAGCCGGATTACCCTACTATATCAAAACACTGTAATCTATGGCGGAATTACCATGATGTCGTGACATCTTGGAGCGCGGTCAAAAACATTATCCAGCACTACGAAGATGAATATTCTGGTTTGGAAAAGTACCACGGACCCGGTCATTGGAATGACCCGGATATGCTAATTTTCGGTACCGATTCCCTGACAGAAGCACAAAGCAAAATTCATATAAGTGTTTACGCAATGCTCTCGGCGCCTTTGCTCTTGAGTTGCGACATGACCCGAGTGAAGGGATACGAAAAGAACCTGCTCTTGAATTTGGACCTCATGGCGATCGCTCGGGATCCGTTGGGGATCATGGCTCGACCTTATAGGATTGATTACATAACTTTATGGGTGAAACCAAGGATGCCGAGGAAGGGCGATATATACCCGTCCTATTCGCTCGCCCTGGTCAATGTCAGCGGTCAGAAACGTACAATAACCGTGAGGCCTTCGAACTACGGCCTCAATTGTACTGACGGGTACAGTGTGATGGACATTTTTAGTAAGAGATTTTTGAAAAATATAACTTTTGAAGATAGCATTGACGTGACCGTTCCTTCTGAAGATGTCATATTATATACTTTGTTCCCGTTGTGA
Protein
MGWMSWGYYMCGADCEQNPDKCLDEKLILSVADTFYNEGYQEAGFEYIIIDDCWSEHHRDRDGRLVPDSLRFPNGMKYISDYIHSRNLKFGMYTNVADVTCMRYQGSKGHFKVDADTFAQWGVDYIKVDGCFVGEQYLNNAYIHFGGYLNKTNRPMVYSCSWPYYIEFIHNKKPDYPTISKHCNLWRNYHDVVTSWSAVKNIIQHYEDEYSGLEKYHGPGHWNDPDMLIFGTDSLTEAQSKIHISVYAMLSAPLLLSCDMTRVKGYEKNLLLNLDLMAIARDPLGIMARPYRIDYITLWVKPRMPRKGDIYPSYSLALVNVSGQKRTITVRPSNYGLNCTDGYSVMDIFSKRFLKNITFEDSIDVTVPSEDVILYTLFPL
Summary
Subunit
Homodimer.
Similarity
Belongs to the glycosyl hydrolase 27 family.
Feature
chain Alpha-galactosidase
Uniprot
H9JWS4
A0A3S2PHR1
A0A0N0P9S8
A0A2A4IW76
A0A2W1BR43
A0A293LAS9
+ More
L7LZY4 A0A131Z618 A0A224YR53 E9HCD9 A0A147BIJ7 D6WTE0 A0A2P8ZLY1 A0A067RL82 A0A0L7QQI1 A0A154PHX5 A0A0P5IJY8 A0A164M2V8 A0A0P5MNZ2 A0A0P5Q8D5 D1ZZP5 A0A0P6ENW6 A0A0P5V6J8 A0A2J7Q6R5 S4NSM8 A0A0P5Q526 A0A3R7M4M2 Q7Q6H3 A0A182V8Y9 A0A182P2V6 A0A1Z5L4A9 Q17AU1 A0A182XMD5 A0A182K837 B4JP37 I4DIL5 A0A0N1PES9 A0A182I269 A0A1B6FU31 A0A182KPZ7 A0A1S4F9T9 A0A1Y1M843 A0A0A1X480 A0A131YVT3 A0A0K2T4T3 A0A0C9R4U7 A0A310SAX6 A0A2W1BQY8 A0A195FQA7 A0A0M8ZS78 E9IEL9 J3JUA1 A0A3M7RVB3 A0A151WST7 D2T1A8 A0A147BG42 A0A2S2PVG7 B4M8H2 A0A034VJ62 A0A026WYH3 A0A182FHK7 A0A0K8WDV6 A0A182H7D4 A0A182T5G1 A0A195BWB5 A0A1A9ZVN6 A0A0P5PTP7 A0A182QQ15 A0A2A4JJP4 A0A2S2PSU3 A0A1A9UT81 V5GMA3 B4KHN3 A0A0P5GIW0 J9JRF8 A0A2J7QEQ5 W5J5I0 A0A0P5HGD4 J9K999 A0A0P6HSF3 A0A2A3EAC4 U4UPP5 A0A067RV68 J9JWZ7 B7PDZ6 A0A0P5P1G6 A0A191UR38 A0A1A9Y6Y0 A0A1A9WLA3 A0A1J1HJ44 A0A0P5CX52 W8B716 E2A7Y1 A0A0P4XF76 A0A0P5S3C2 A0A0P4XZR8 A0A182WLA6 A0A0P5WG04 A0A0P5DDU7
L7LZY4 A0A131Z618 A0A224YR53 E9HCD9 A0A147BIJ7 D6WTE0 A0A2P8ZLY1 A0A067RL82 A0A0L7QQI1 A0A154PHX5 A0A0P5IJY8 A0A164M2V8 A0A0P5MNZ2 A0A0P5Q8D5 D1ZZP5 A0A0P6ENW6 A0A0P5V6J8 A0A2J7Q6R5 S4NSM8 A0A0P5Q526 A0A3R7M4M2 Q7Q6H3 A0A182V8Y9 A0A182P2V6 A0A1Z5L4A9 Q17AU1 A0A182XMD5 A0A182K837 B4JP37 I4DIL5 A0A0N1PES9 A0A182I269 A0A1B6FU31 A0A182KPZ7 A0A1S4F9T9 A0A1Y1M843 A0A0A1X480 A0A131YVT3 A0A0K2T4T3 A0A0C9R4U7 A0A310SAX6 A0A2W1BQY8 A0A195FQA7 A0A0M8ZS78 E9IEL9 J3JUA1 A0A3M7RVB3 A0A151WST7 D2T1A8 A0A147BG42 A0A2S2PVG7 B4M8H2 A0A034VJ62 A0A026WYH3 A0A182FHK7 A0A0K8WDV6 A0A182H7D4 A0A182T5G1 A0A195BWB5 A0A1A9ZVN6 A0A0P5PTP7 A0A182QQ15 A0A2A4JJP4 A0A2S2PSU3 A0A1A9UT81 V5GMA3 B4KHN3 A0A0P5GIW0 J9JRF8 A0A2J7QEQ5 W5J5I0 A0A0P5HGD4 J9K999 A0A0P6HSF3 A0A2A3EAC4 U4UPP5 A0A067RV68 J9JWZ7 B7PDZ6 A0A0P5P1G6 A0A191UR38 A0A1A9Y6Y0 A0A1A9WLA3 A0A1J1HJ44 A0A0P5CX52 W8B716 E2A7Y1 A0A0P4XF76 A0A0P5S3C2 A0A0P4XZR8 A0A182WLA6 A0A0P5WG04 A0A0P5DDU7
EC Number
3.2.1.-
Pubmed
19121390
26354079
28756777
25576852
26830274
28797301
+ More
21292972 29652888 18362917 19820115 29403074 24845553 23622113 12364791 14747013 17210077 28528879 17510324 17994087 22651552 20966253 28004739 25830018 21282665 22516182 30375419 25348373 24508170 26483478 20920257 23761445 23537049 27142481 24495485 20798317
21292972 29652888 18362917 19820115 29403074 24845553 23622113 12364791 14747013 17210077 28528879 17510324 17994087 22651552 20966253 28004739 25830018 21282665 22516182 30375419 25348373 24508170 26483478 20920257 23761445 23537049 27142481 24495485 20798317
EMBL
BABH01018570
RSAL01000029
RVE51771.1
KQ459166
KPJ03548.1
NWSH01006121
+ More
PCG63648.1 KZ149970 PZC76085.1 GFWV01000121 MAA24851.1 GACK01008581 JAA56453.1 GEDV01002139 JAP86418.1 GFPF01005925 MAA17071.1 GL732619 EFX70604.1 GEGO01005279 JAR90125.1 KQ971352 EFA06705.2 PYGN01000019 PSN57511.1 KK852444 KDR23773.1 KQ414786 KOC60877.1 KQ434896 KZC10800.1 GDIQ01212410 JAK39315.1 LRGB01003101 KZS04676.1 GDIQ01153464 JAK98261.1 GDIQ01139344 JAL12382.1 KQ971338 EFA01818.1 GDIQ01060719 JAN34018.1 GDIP01119606 GDIP01079951 JAL84108.1 NEVH01017454 PNF24276.1 GAIX01014012 JAA78548.1 GDIQ01119290 JAL32436.1 QCYY01002113 ROT72774.1 AAAB01008960 EAA11949.3 GFJQ02004737 JAW02233.1 CH477330 EAT43367.1 CH916372 EDV99462.1 AK401133 BAM17755.1 KQ458751 KPJ05313.1 APCN01000126 GECZ01016138 GECZ01015544 JAS53631.1 JAS54225.1 GEZM01037887 JAV81864.1 GBXI01008395 JAD05897.1 GEDV01005905 JAP82652.1 HACA01003085 CDW20446.1 GBYB01007777 JAG77544.1 KQ765605 OAD53851.1 KZ149927 PZC77532.1 KQ981335 KYN42658.1 KQ435878 KOX70065.1 GL762647 EFZ20978.1 BT126814 AEE61776.1 REGN01002531 RNA27511.1 KQ982769 KYQ50857.1 FM986353 CAX20733.1 GEGO01005666 JAR89738.1 GGMS01000258 MBY69461.1 CH940654 EDW57498.2 GAKP01016413 JAC42539.1 KK107079 EZA60164.1 GDHF01006436 GDHF01002966 JAI45878.1 JAI49348.1 JXUM01028208 JXUM01028209 KQ560814 KXJ80784.1 KQ976401 KYM92575.1 GDIQ01133997 JAL17729.1 AXCN02000446 NWSH01001168 PCG72287.1 GGMR01019932 MBY32551.1 GALX01005719 JAB62747.1 CH933807 EDW12312.1 GDIQ01246250 JAK05475.1 ABLF02027051 ABLF02027053 NEVH01015307 PNF27047.1 ADMH02002096 ETN59246.1 GDIQ01233405 JAK18320.1 ABLF02030053 GDIQ01022778 JAN71959.1 KZ288322 PBC28159.1 KB632308 ERL91995.1 KDR23774.1 ABLF02027580 ABJB010864680 ABJB010932589 ABJB011004459 DS692880 EEC04818.1 GDIQ01136536 JAL15190.1 KU365924 ANJ04640.1 CVRI01000001 CRK86297.1 GDIP01164198 JAJ59204.1 GAMC01009600 JAB96955.1 GL437442 EFN70460.1 GDIP01251680 JAI71721.1 GDIQ01093583 JAL58143.1 GDIP01234586 JAI88815.1 GDIP01215792 GDIP01099894 LRGB01003056 JAM03821.1 KZS04814.1 GDIP01158942 JAJ64460.1
PCG63648.1 KZ149970 PZC76085.1 GFWV01000121 MAA24851.1 GACK01008581 JAA56453.1 GEDV01002139 JAP86418.1 GFPF01005925 MAA17071.1 GL732619 EFX70604.1 GEGO01005279 JAR90125.1 KQ971352 EFA06705.2 PYGN01000019 PSN57511.1 KK852444 KDR23773.1 KQ414786 KOC60877.1 KQ434896 KZC10800.1 GDIQ01212410 JAK39315.1 LRGB01003101 KZS04676.1 GDIQ01153464 JAK98261.1 GDIQ01139344 JAL12382.1 KQ971338 EFA01818.1 GDIQ01060719 JAN34018.1 GDIP01119606 GDIP01079951 JAL84108.1 NEVH01017454 PNF24276.1 GAIX01014012 JAA78548.1 GDIQ01119290 JAL32436.1 QCYY01002113 ROT72774.1 AAAB01008960 EAA11949.3 GFJQ02004737 JAW02233.1 CH477330 EAT43367.1 CH916372 EDV99462.1 AK401133 BAM17755.1 KQ458751 KPJ05313.1 APCN01000126 GECZ01016138 GECZ01015544 JAS53631.1 JAS54225.1 GEZM01037887 JAV81864.1 GBXI01008395 JAD05897.1 GEDV01005905 JAP82652.1 HACA01003085 CDW20446.1 GBYB01007777 JAG77544.1 KQ765605 OAD53851.1 KZ149927 PZC77532.1 KQ981335 KYN42658.1 KQ435878 KOX70065.1 GL762647 EFZ20978.1 BT126814 AEE61776.1 REGN01002531 RNA27511.1 KQ982769 KYQ50857.1 FM986353 CAX20733.1 GEGO01005666 JAR89738.1 GGMS01000258 MBY69461.1 CH940654 EDW57498.2 GAKP01016413 JAC42539.1 KK107079 EZA60164.1 GDHF01006436 GDHF01002966 JAI45878.1 JAI49348.1 JXUM01028208 JXUM01028209 KQ560814 KXJ80784.1 KQ976401 KYM92575.1 GDIQ01133997 JAL17729.1 AXCN02000446 NWSH01001168 PCG72287.1 GGMR01019932 MBY32551.1 GALX01005719 JAB62747.1 CH933807 EDW12312.1 GDIQ01246250 JAK05475.1 ABLF02027051 ABLF02027053 NEVH01015307 PNF27047.1 ADMH02002096 ETN59246.1 GDIQ01233405 JAK18320.1 ABLF02030053 GDIQ01022778 JAN71959.1 KZ288322 PBC28159.1 KB632308 ERL91995.1 KDR23774.1 ABLF02027580 ABJB010864680 ABJB010932589 ABJB011004459 DS692880 EEC04818.1 GDIQ01136536 JAL15190.1 KU365924 ANJ04640.1 CVRI01000001 CRK86297.1 GDIP01164198 JAJ59204.1 GAMC01009600 JAB96955.1 GL437442 EFN70460.1 GDIP01251680 JAI71721.1 GDIQ01093583 JAL58143.1 GDIP01234586 JAI88815.1 GDIP01215792 GDIP01099894 LRGB01003056 JAM03821.1 KZS04814.1 GDIP01158942 JAJ64460.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000218220
UP000000305
UP000007266
+ More
UP000245037 UP000027135 UP000053825 UP000076502 UP000076858 UP000235965 UP000283509 UP000007062 UP000075903 UP000075885 UP000008820 UP000076407 UP000075881 UP000001070 UP000075840 UP000075882 UP000078541 UP000053105 UP000276133 UP000075809 UP000008792 UP000053097 UP000069272 UP000069940 UP000249989 UP000075901 UP000078540 UP000092445 UP000075886 UP000078200 UP000009192 UP000007819 UP000000673 UP000242457 UP000030742 UP000001555 UP000092443 UP000091820 UP000183832 UP000000311 UP000075920
UP000245037 UP000027135 UP000053825 UP000076502 UP000076858 UP000235965 UP000283509 UP000007062 UP000075903 UP000075885 UP000008820 UP000076407 UP000075881 UP000001070 UP000075840 UP000075882 UP000078541 UP000053105 UP000276133 UP000075809 UP000008792 UP000053097 UP000069272 UP000069940 UP000249989 UP000075901 UP000078540 UP000092445 UP000075886 UP000078200 UP000009192 UP000007819 UP000000673 UP000242457 UP000030742 UP000001555 UP000092443 UP000091820 UP000183832 UP000000311 UP000075920
PRIDE
Interpro
SUPFAM
SSF51445
SSF51445
Gene 3D
CDD
ProteinModelPortal
H9JWS4
A0A3S2PHR1
A0A0N0P9S8
A0A2A4IW76
A0A2W1BR43
A0A293LAS9
+ More
L7LZY4 A0A131Z618 A0A224YR53 E9HCD9 A0A147BIJ7 D6WTE0 A0A2P8ZLY1 A0A067RL82 A0A0L7QQI1 A0A154PHX5 A0A0P5IJY8 A0A164M2V8 A0A0P5MNZ2 A0A0P5Q8D5 D1ZZP5 A0A0P6ENW6 A0A0P5V6J8 A0A2J7Q6R5 S4NSM8 A0A0P5Q526 A0A3R7M4M2 Q7Q6H3 A0A182V8Y9 A0A182P2V6 A0A1Z5L4A9 Q17AU1 A0A182XMD5 A0A182K837 B4JP37 I4DIL5 A0A0N1PES9 A0A182I269 A0A1B6FU31 A0A182KPZ7 A0A1S4F9T9 A0A1Y1M843 A0A0A1X480 A0A131YVT3 A0A0K2T4T3 A0A0C9R4U7 A0A310SAX6 A0A2W1BQY8 A0A195FQA7 A0A0M8ZS78 E9IEL9 J3JUA1 A0A3M7RVB3 A0A151WST7 D2T1A8 A0A147BG42 A0A2S2PVG7 B4M8H2 A0A034VJ62 A0A026WYH3 A0A182FHK7 A0A0K8WDV6 A0A182H7D4 A0A182T5G1 A0A195BWB5 A0A1A9ZVN6 A0A0P5PTP7 A0A182QQ15 A0A2A4JJP4 A0A2S2PSU3 A0A1A9UT81 V5GMA3 B4KHN3 A0A0P5GIW0 J9JRF8 A0A2J7QEQ5 W5J5I0 A0A0P5HGD4 J9K999 A0A0P6HSF3 A0A2A3EAC4 U4UPP5 A0A067RV68 J9JWZ7 B7PDZ6 A0A0P5P1G6 A0A191UR38 A0A1A9Y6Y0 A0A1A9WLA3 A0A1J1HJ44 A0A0P5CX52 W8B716 E2A7Y1 A0A0P4XF76 A0A0P5S3C2 A0A0P4XZR8 A0A182WLA6 A0A0P5WG04 A0A0P5DDU7
L7LZY4 A0A131Z618 A0A224YR53 E9HCD9 A0A147BIJ7 D6WTE0 A0A2P8ZLY1 A0A067RL82 A0A0L7QQI1 A0A154PHX5 A0A0P5IJY8 A0A164M2V8 A0A0P5MNZ2 A0A0P5Q8D5 D1ZZP5 A0A0P6ENW6 A0A0P5V6J8 A0A2J7Q6R5 S4NSM8 A0A0P5Q526 A0A3R7M4M2 Q7Q6H3 A0A182V8Y9 A0A182P2V6 A0A1Z5L4A9 Q17AU1 A0A182XMD5 A0A182K837 B4JP37 I4DIL5 A0A0N1PES9 A0A182I269 A0A1B6FU31 A0A182KPZ7 A0A1S4F9T9 A0A1Y1M843 A0A0A1X480 A0A131YVT3 A0A0K2T4T3 A0A0C9R4U7 A0A310SAX6 A0A2W1BQY8 A0A195FQA7 A0A0M8ZS78 E9IEL9 J3JUA1 A0A3M7RVB3 A0A151WST7 D2T1A8 A0A147BG42 A0A2S2PVG7 B4M8H2 A0A034VJ62 A0A026WYH3 A0A182FHK7 A0A0K8WDV6 A0A182H7D4 A0A182T5G1 A0A195BWB5 A0A1A9ZVN6 A0A0P5PTP7 A0A182QQ15 A0A2A4JJP4 A0A2S2PSU3 A0A1A9UT81 V5GMA3 B4KHN3 A0A0P5GIW0 J9JRF8 A0A2J7QEQ5 W5J5I0 A0A0P5HGD4 J9K999 A0A0P6HSF3 A0A2A3EAC4 U4UPP5 A0A067RV68 J9JWZ7 B7PDZ6 A0A0P5P1G6 A0A191UR38 A0A1A9Y6Y0 A0A1A9WLA3 A0A1J1HJ44 A0A0P5CX52 W8B716 E2A7Y1 A0A0P4XF76 A0A0P5S3C2 A0A0P4XZR8 A0A182WLA6 A0A0P5WG04 A0A0P5DDU7
PDB
1KTC
E-value=4.31122e-75,
Score=715
Ontologies
PATHWAY
GO
PANTHER
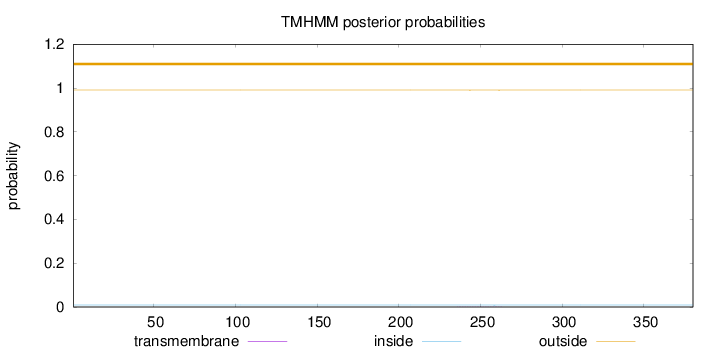
Topology
Length:
380
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01793
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00877
outside
1 - 380
Population Genetic Test Statistics
Pi
191.609186
Theta
166.449978
Tajima's D
0.941335
CLR
0.082838
CSRT
0.644117794110294
Interpretation
Uncertain
