Gene
KWMTBOMO16239
Pre Gene Modal
BGIBMGA013908
Annotation
PREDICTED:_putative_ferric-chelate_reductase_1_homolog_[Amyelois_transitella]
Full name
Putative ferric-chelate reductase 1 homolog
Location in the cell
PlasmaMembrane Reliability : 4.674
Sequence
CDS
ATGTTGCCAAGAAAAAGGAATATCATCTTCGCGCTGCTCGTTTTGGTTCCGTTTTCTGAACAGTACAGTTCCGGAGCTCCTCCTGGAGCCTGCGAAGATCAGCTGCCTCGTCACGAATCTATTGTACCACAATCGTCGGCGCCTCCATACTTAATACTGACGTCCAGTGCTCAAGTCAGGCAAGGCGACACGCTCAACGTCACCATCGGAAGTCCCGCCGGCGCCCCTGTGCCTATCGGTGGGTTCTTGCTCCAAGCTAGAGAAATACAGAATCCAGATGCGATTGTCGGTAAATTTGTCTCGGTTCCCGCCACTGAAACGCACGTGACGACATGCGCCAAGCCGAATGATACGGTCACGCATTCGTCACCCGCGGACAAAGGAGCTTTATCGTTCCGGTGGCAAGCCCCAACTGATTTCTTAGGAGGCGTTGAATTCAGAGCGACCGTAGCACAGAGCTACGCCACATTCTGGAAGGGCGTCGAATCTCCCCTAGTCGAAGTGGTCACTCCGAACACGGAAATCACGACGCAAATGGCACAATCTACTTCTACAACTACCGCTCGTGCACCGCCTGTGATTATGGAAAATAAGCCGAAGCTAACGACCCAAAAACTAGACGAAATCTACCAAGGCTGTGCCGACACTAAACTGTGTTTTGGCGTACCCCAAAACTGCATAGAGAAAGGCGATTGCAAAGCCATTGTGGCCGTATTCGTCGCGGGAGACGTTTACACATTCGAAGTCCAAGGCACTGGGAATCCGAAATACGTGGCGGCTGCTCTGAGTTTGGACACTAAAATGGGCGACGACAGCGCTATGGAGTGCGTCAGGAACAACAATGGCATCATTAATTTATACACCTCCTGGACATACCCGAAAGTGGAGCCCTATGTTAATAGATCAGATTCTCCACAAGATATAGTGCAGTTGCTCCAATCTTCGACGATCGATGGAAAACTTTATTGTAAGTTCACGAGAGACGCGGTCTCGACTGTCAAGGGAATAAAGTTTGATCTGATGAACAAAAAATACAATCTGATGATTGTCTCCGGTGACGGCATGAAAGATCCAGAACGCGTAGGCTTCCATACGATAGCTTATGAATCATCAGGCGAACCGTTGGCGCTGTCCAGTGTCGGAACAGCGGCTGGCGCTTCCAAACTGTTGCTCAAACTGCACGGCTGCTTCATGGTGGCCGCCTGGCTGGGAGCCTCGTCCCTGGGAATCATACTGGCCAGGTACTACAGGCAGACCTGGGTGGGAAGGCAGCTCGGCGGGAAGGACATATGGTTTGCCTATCATCGTATCTTCATGATGACCACGTGGCTGCTGACAATGGCCGGTTTCGTCCTGATCCTAATTGAGGCTAGAGGCTGGTCCGACACCGGAGACAACCCTCACGCCATCACCGGGATCATAACGGTTGTTCTCTGCTTCTTACAGCCCATTGGTGCATTCTTCAGACCTCACCCCGGTACTAATAAGAGGATCTACTTCAACTGGACGCATTGGCTCGCCGGGAATACGGCTCACATCTTGGGCATTGTAACAATATTCTTCGCGGTGTACCTCCAAAAGGCGGAATTACCGGCGTGGACCACCTTCGTCCTGGCAGCCTTCGTGGCATTCCACGTGGTCATGCACCTCGTACTAACGTTGACAGCCTGTGTATCGGACGGCAGTATAGGTTCTGCGCGAGTCAACGCCTTCCCTATGAAAGACATGCTCGGTGGCACCCGACCCACGCAGATAGATCGGACTGGAGATGCACCGTATTCCGGCTTCAGAAGATTACTTTTAGGTATATACGGGCCCGTTGTACTGCTGTTCGCGCTGGCTGTCGTATGTCTGGTAGCTTTGGCGCCAATCACAGAAACTTATAACAATATTATGGGAGCCTAA
Protein
MLPRKRNIIFALLVLVPFSEQYSSGAPPGACEDQLPRHESIVPQSSAPPYLILTSSAQVRQGDTLNVTIGSPAGAPVPIGGFLLQAREIQNPDAIVGKFVSVPATETHVTTCAKPNDTVTHSSPADKGALSFRWQAPTDFLGGVEFRATVAQSYATFWKGVESPLVEVVTPNTEITTQMAQSTSTTTARAPPVIMENKPKLTTQKLDEIYQGCADTKLCFGVPQNCIEKGDCKAIVAVFVAGDVYTFEVQGTGNPKYVAAALSLDTKMGDDSAMECVRNNNGIINLYTSWTYPKVEPYVNRSDSPQDIVQLLQSSTIDGKLYCKFTRDAVSTVKGIKFDLMNKKYNLMIVSGDGMKDPERVGFHTIAYESSGEPLALSSVGTAAGASKLLLKLHGCFMVAAWLGASSLGIILARYYRQTWVGRQLGGKDIWFAYHRIFMMTTWLLTMAGFVLILIEARGWSDTGDNPHAITGIITVVLCFLQPIGAFFRPHPGTNKRIYFNWTHWLAGNTAHILGIVTIFFAVYLQKAELPAWTTFVLAAFVAFHVVMHLVLTLTACVSDGSIGSARVNAFPMKDMLGGTRPTQIDRTGDAPYSGFRRLLLGIYGPVVLLFALAVVCLVALAPITETYNNIMGA
Summary
Description
Putative ferric-chelate reductases reduce Fe(3+) to Fe(2+) before its transport from the endosome to the cytoplasm.
Similarity
Belongs to the FRRS1 family.
Keywords
Complete proteome
Electron transport
Glycoprotein
Membrane
Oxidoreductase
Reference proteome
Transmembrane
Transmembrane helix
Transport
Feature
chain Putative ferric-chelate reductase 1 homolog
Uniprot
H9JWJ4
A0A2A4JFI9
A0A2W1BM05
A0A0N0P9S7
A0A2H1WPB9
A0A212FF09
+ More
I4DNL5 A0A0N1IGU8 A0A0L7LP55 A0A1Q3F062 A0A1Q3F146 A0A084W2B7 B0WE73 A0A2M3Z0A1 A0A1Q3F140 A0A182IJ90 A0A336KI37 A0A336KXK1 A0A1Q3F069 A0A182F9M2 W5JCW7 A0A336KZF5 A0A2M4BH83 A0A2M4A6S7 A0A2M4A817 A0A2M4BH51 A0A336KMH5 A0A336KNF6 A0A336L231 Q7Q9L3 A0A2M4A6W7 Q176N6 A0A182R3E9 U5EVK0 A0A182ST67 A0A182M4V6 Q176N7 A0A023EXF7 A0A1A9WDX6 A0A182YIG5 A0A0A1WJY4 A0A182KFZ1 A0A0K8WG94 A0A182N1N9 A0A182V3C9 A0A1Q3EZY2 D6WU73 A0A034VR07 A0A0K8TYT4 A0A182TQ49 A0A034VSM5 A0A182PCI4 E0VAI3 A0A0K8WMI5 W8C6I9 A0A023EV28 A0A182Q8D5 A0A182H6L9 A0A023EV19 A0A1L8E328 A0A0J9RDZ5 A0A1L8E3J5 A0A182HP18 B4QHE1 A0A182WAF8 A0A1B0FPS2 B4P6S3 A0A1Y1NJQ2 A0A1I8Q687 A0A182XEZ2 A0A0B4LGL5 A0A1A9UI54 Q8MSU3 B4LL45 B3NPY4 A0A0Q9W5G4 A0A2J7Q8D6 B3MF59 A0A0M4E532 A0A232F001 B4KQB9 A0A0Q9X840 A0A1J1IIG7 A0A3B0IZZ9 A0A1W4VDZ3 B4J9X2 B4GG47 T1PEE9 E1ZX58 A0A1I8MVF4 A0A3B0IZQ1 N6WCS6 A0A1A9Z5K9 K7J493 A0A1L8DJP5 A0A151IHA2 A0A0L0CBB6 A0A1I8MVF8 Q28XH2 A0A0N0U5T5
I4DNL5 A0A0N1IGU8 A0A0L7LP55 A0A1Q3F062 A0A1Q3F146 A0A084W2B7 B0WE73 A0A2M3Z0A1 A0A1Q3F140 A0A182IJ90 A0A336KI37 A0A336KXK1 A0A1Q3F069 A0A182F9M2 W5JCW7 A0A336KZF5 A0A2M4BH83 A0A2M4A6S7 A0A2M4A817 A0A2M4BH51 A0A336KMH5 A0A336KNF6 A0A336L231 Q7Q9L3 A0A2M4A6W7 Q176N6 A0A182R3E9 U5EVK0 A0A182ST67 A0A182M4V6 Q176N7 A0A023EXF7 A0A1A9WDX6 A0A182YIG5 A0A0A1WJY4 A0A182KFZ1 A0A0K8WG94 A0A182N1N9 A0A182V3C9 A0A1Q3EZY2 D6WU73 A0A034VR07 A0A0K8TYT4 A0A182TQ49 A0A034VSM5 A0A182PCI4 E0VAI3 A0A0K8WMI5 W8C6I9 A0A023EV28 A0A182Q8D5 A0A182H6L9 A0A023EV19 A0A1L8E328 A0A0J9RDZ5 A0A1L8E3J5 A0A182HP18 B4QHE1 A0A182WAF8 A0A1B0FPS2 B4P6S3 A0A1Y1NJQ2 A0A1I8Q687 A0A182XEZ2 A0A0B4LGL5 A0A1A9UI54 Q8MSU3 B4LL45 B3NPY4 A0A0Q9W5G4 A0A2J7Q8D6 B3MF59 A0A0M4E532 A0A232F001 B4KQB9 A0A0Q9X840 A0A1J1IIG7 A0A3B0IZZ9 A0A1W4VDZ3 B4J9X2 B4GG47 T1PEE9 E1ZX58 A0A1I8MVF4 A0A3B0IZQ1 N6WCS6 A0A1A9Z5K9 K7J493 A0A1L8DJP5 A0A151IHA2 A0A0L0CBB6 A0A1I8MVF8 Q28XH2 A0A0N0U5T5
EC Number
1.-.-.-
Pubmed
19121390
28756777
26354079
22118469
22651552
26227816
+ More
24438588 20920257 23761445 12364791 14747013 17210077 17510324 24945155 26483478 25244985 25830018 18362917 19820115 25348373 20566863 24495485 22936249 17994087 17550304 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 14499595 18057021 28648823 20798317 25315136 15632085 20075255 26108605
24438588 20920257 23761445 12364791 14747013 17210077 17510324 24945155 26483478 25244985 25830018 18362917 19820115 25348373 20566863 24495485 22936249 17994087 17550304 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 14499595 18057021 28648823 20798317 25315136 15632085 20075255 26108605
EMBL
BABH01018561
BABH01018562
BABH01018563
BABH01018564
BABH01018565
NWSH01001563
+ More
PCG70855.1 KZ149970 PZC76089.1 KQ459166 KPJ03545.1 ODYU01010037 SOQ54857.1 AGBW02008868 OWR52331.1 AK402994 BAM19505.1 KQ460689 KPJ12745.1 JTDY01000478 KOB77001.1 GFDL01014103 JAV20942.1 GFDL01013781 JAV21264.1 ATLV01019596 KE525275 KFB44361.1 DS231905 EDS45300.1 GGFM01001211 MBW21962.1 GFDL01013791 JAV21254.1 UFQS01000514 UFQT01000514 SSX04524.1 SSX24887.1 UFQS01001309 UFQT01001309 SSX10252.1 SSX29940.1 GFDL01014099 JAV20946.1 ADMH02001867 ETN60735.1 SSX10250.1 SSX29938.1 GGFJ01003243 MBW52384.1 GGFK01003182 MBW36503.1 GGFK01003584 MBW36905.1 GGFJ01003244 MBW52385.1 SSX04525.1 SSX24888.1 SSX04523.1 SSX24886.1 SSX10251.1 SSX29939.1 AAAB01008900 EAA09429.4 GGFK01003225 MBW36546.1 CH477385 EAT42110.1 GANO01001827 JAB58044.1 AXCM01004801 EAT42111.1 JXUM01112707 JXUM01112708 GAPW01000489 KQ565616 JAC13109.1 KXJ70842.1 GBXI01015497 JAC98794.1 GDHF01002245 GDHF01000676 JAI50069.1 JAI51638.1 GFDL01014191 JAV20854.1 KQ971352 EFA06259.1 GAKP01014395 JAC44557.1 GDHF01032873 GDHF01024606 JAI19441.1 JAI27708.1 GAKP01014394 JAC44558.1 DS235006 EEB10389.1 GDHF01000214 JAI52100.1 GAMC01008506 GAMC01008504 GAMC01008501 GAMC01008500 JAB98054.1 GAPW01000736 JAC12862.1 AXCN02000999 JXUM01027162 KQ560781 KXJ80916.1 GAPW01000746 JAC12852.1 GFDF01000941 JAV13143.1 CM002911 KMY94146.1 GFDF01000942 JAV13142.1 APCN01002869 CM000362 EDX07286.1 KMY94145.1 CCAG010015309 CM000158 EDW92000.1 GEZM01005773 GEZM01005772 JAV95927.1 AE013599 AHN56269.1 AY118595 CH940648 EDW61852.1 CH954179 EDV55831.1 KRF80283.1 KRF80284.1 NEVH01016959 PNF24839.1 CH902619 EDV37687.1 KPU76989.1 CP012524 ALC41304.1 NNAY01001471 OXU23850.1 CH933808 EDW09247.2 KRG04543.1 CVRI01000054 CRL00019.1 OUUW01000001 SPP73935.1 CH916367 EDW02559.1 CH479183 EDW35467.1 KA646318 AFP60947.1 GL435030 EFN74148.1 SPP73934.1 CM000071 ENO01889.1 AAZX01003246 GFDF01007385 JAV06699.1 KQ977636 KYN01150.1 JRES01000655 KNC29551.1 EAL26344.3 KQ435756 KOX75901.1
PCG70855.1 KZ149970 PZC76089.1 KQ459166 KPJ03545.1 ODYU01010037 SOQ54857.1 AGBW02008868 OWR52331.1 AK402994 BAM19505.1 KQ460689 KPJ12745.1 JTDY01000478 KOB77001.1 GFDL01014103 JAV20942.1 GFDL01013781 JAV21264.1 ATLV01019596 KE525275 KFB44361.1 DS231905 EDS45300.1 GGFM01001211 MBW21962.1 GFDL01013791 JAV21254.1 UFQS01000514 UFQT01000514 SSX04524.1 SSX24887.1 UFQS01001309 UFQT01001309 SSX10252.1 SSX29940.1 GFDL01014099 JAV20946.1 ADMH02001867 ETN60735.1 SSX10250.1 SSX29938.1 GGFJ01003243 MBW52384.1 GGFK01003182 MBW36503.1 GGFK01003584 MBW36905.1 GGFJ01003244 MBW52385.1 SSX04525.1 SSX24888.1 SSX04523.1 SSX24886.1 SSX10251.1 SSX29939.1 AAAB01008900 EAA09429.4 GGFK01003225 MBW36546.1 CH477385 EAT42110.1 GANO01001827 JAB58044.1 AXCM01004801 EAT42111.1 JXUM01112707 JXUM01112708 GAPW01000489 KQ565616 JAC13109.1 KXJ70842.1 GBXI01015497 JAC98794.1 GDHF01002245 GDHF01000676 JAI50069.1 JAI51638.1 GFDL01014191 JAV20854.1 KQ971352 EFA06259.1 GAKP01014395 JAC44557.1 GDHF01032873 GDHF01024606 JAI19441.1 JAI27708.1 GAKP01014394 JAC44558.1 DS235006 EEB10389.1 GDHF01000214 JAI52100.1 GAMC01008506 GAMC01008504 GAMC01008501 GAMC01008500 JAB98054.1 GAPW01000736 JAC12862.1 AXCN02000999 JXUM01027162 KQ560781 KXJ80916.1 GAPW01000746 JAC12852.1 GFDF01000941 JAV13143.1 CM002911 KMY94146.1 GFDF01000942 JAV13142.1 APCN01002869 CM000362 EDX07286.1 KMY94145.1 CCAG010015309 CM000158 EDW92000.1 GEZM01005773 GEZM01005772 JAV95927.1 AE013599 AHN56269.1 AY118595 CH940648 EDW61852.1 CH954179 EDV55831.1 KRF80283.1 KRF80284.1 NEVH01016959 PNF24839.1 CH902619 EDV37687.1 KPU76989.1 CP012524 ALC41304.1 NNAY01001471 OXU23850.1 CH933808 EDW09247.2 KRG04543.1 CVRI01000054 CRL00019.1 OUUW01000001 SPP73935.1 CH916367 EDW02559.1 CH479183 EDW35467.1 KA646318 AFP60947.1 GL435030 EFN74148.1 SPP73934.1 CM000071 ENO01889.1 AAZX01003246 GFDF01007385 JAV06699.1 KQ977636 KYN01150.1 JRES01000655 KNC29551.1 EAL26344.3 KQ435756 KOX75901.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000037510
+ More
UP000030765 UP000002320 UP000075880 UP000069272 UP000000673 UP000007062 UP000008820 UP000075900 UP000075901 UP000075883 UP000069940 UP000249989 UP000091820 UP000076408 UP000075881 UP000075884 UP000075903 UP000007266 UP000075902 UP000075885 UP000009046 UP000075886 UP000075840 UP000000304 UP000075920 UP000092444 UP000002282 UP000095300 UP000076407 UP000000803 UP000078200 UP000008792 UP000008711 UP000235965 UP000007801 UP000092553 UP000215335 UP000009192 UP000183832 UP000268350 UP000192221 UP000001070 UP000008744 UP000000311 UP000095301 UP000001819 UP000092445 UP000002358 UP000078542 UP000037069 UP000053105
UP000030765 UP000002320 UP000075880 UP000069272 UP000000673 UP000007062 UP000008820 UP000075900 UP000075901 UP000075883 UP000069940 UP000249989 UP000091820 UP000076408 UP000075881 UP000075884 UP000075903 UP000007266 UP000075902 UP000075885 UP000009046 UP000075886 UP000075840 UP000000304 UP000075920 UP000092444 UP000002282 UP000095300 UP000076407 UP000000803 UP000078200 UP000008792 UP000008711 UP000235965 UP000007801 UP000092553 UP000215335 UP000009192 UP000183832 UP000268350 UP000192221 UP000001070 UP000008744 UP000000311 UP000095301 UP000001819 UP000092445 UP000002358 UP000078542 UP000037069 UP000053105
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JWJ4
A0A2A4JFI9
A0A2W1BM05
A0A0N0P9S7
A0A2H1WPB9
A0A212FF09
+ More
I4DNL5 A0A0N1IGU8 A0A0L7LP55 A0A1Q3F062 A0A1Q3F146 A0A084W2B7 B0WE73 A0A2M3Z0A1 A0A1Q3F140 A0A182IJ90 A0A336KI37 A0A336KXK1 A0A1Q3F069 A0A182F9M2 W5JCW7 A0A336KZF5 A0A2M4BH83 A0A2M4A6S7 A0A2M4A817 A0A2M4BH51 A0A336KMH5 A0A336KNF6 A0A336L231 Q7Q9L3 A0A2M4A6W7 Q176N6 A0A182R3E9 U5EVK0 A0A182ST67 A0A182M4V6 Q176N7 A0A023EXF7 A0A1A9WDX6 A0A182YIG5 A0A0A1WJY4 A0A182KFZ1 A0A0K8WG94 A0A182N1N9 A0A182V3C9 A0A1Q3EZY2 D6WU73 A0A034VR07 A0A0K8TYT4 A0A182TQ49 A0A034VSM5 A0A182PCI4 E0VAI3 A0A0K8WMI5 W8C6I9 A0A023EV28 A0A182Q8D5 A0A182H6L9 A0A023EV19 A0A1L8E328 A0A0J9RDZ5 A0A1L8E3J5 A0A182HP18 B4QHE1 A0A182WAF8 A0A1B0FPS2 B4P6S3 A0A1Y1NJQ2 A0A1I8Q687 A0A182XEZ2 A0A0B4LGL5 A0A1A9UI54 Q8MSU3 B4LL45 B3NPY4 A0A0Q9W5G4 A0A2J7Q8D6 B3MF59 A0A0M4E532 A0A232F001 B4KQB9 A0A0Q9X840 A0A1J1IIG7 A0A3B0IZZ9 A0A1W4VDZ3 B4J9X2 B4GG47 T1PEE9 E1ZX58 A0A1I8MVF4 A0A3B0IZQ1 N6WCS6 A0A1A9Z5K9 K7J493 A0A1L8DJP5 A0A151IHA2 A0A0L0CBB6 A0A1I8MVF8 Q28XH2 A0A0N0U5T5
I4DNL5 A0A0N1IGU8 A0A0L7LP55 A0A1Q3F062 A0A1Q3F146 A0A084W2B7 B0WE73 A0A2M3Z0A1 A0A1Q3F140 A0A182IJ90 A0A336KI37 A0A336KXK1 A0A1Q3F069 A0A182F9M2 W5JCW7 A0A336KZF5 A0A2M4BH83 A0A2M4A6S7 A0A2M4A817 A0A2M4BH51 A0A336KMH5 A0A336KNF6 A0A336L231 Q7Q9L3 A0A2M4A6W7 Q176N6 A0A182R3E9 U5EVK0 A0A182ST67 A0A182M4V6 Q176N7 A0A023EXF7 A0A1A9WDX6 A0A182YIG5 A0A0A1WJY4 A0A182KFZ1 A0A0K8WG94 A0A182N1N9 A0A182V3C9 A0A1Q3EZY2 D6WU73 A0A034VR07 A0A0K8TYT4 A0A182TQ49 A0A034VSM5 A0A182PCI4 E0VAI3 A0A0K8WMI5 W8C6I9 A0A023EV28 A0A182Q8D5 A0A182H6L9 A0A023EV19 A0A1L8E328 A0A0J9RDZ5 A0A1L8E3J5 A0A182HP18 B4QHE1 A0A182WAF8 A0A1B0FPS2 B4P6S3 A0A1Y1NJQ2 A0A1I8Q687 A0A182XEZ2 A0A0B4LGL5 A0A1A9UI54 Q8MSU3 B4LL45 B3NPY4 A0A0Q9W5G4 A0A2J7Q8D6 B3MF59 A0A0M4E532 A0A232F001 B4KQB9 A0A0Q9X840 A0A1J1IIG7 A0A3B0IZZ9 A0A1W4VDZ3 B4J9X2 B4GG47 T1PEE9 E1ZX58 A0A1I8MVF4 A0A3B0IZQ1 N6WCS6 A0A1A9Z5K9 K7J493 A0A1L8DJP5 A0A151IHA2 A0A0L0CBB6 A0A1I8MVF8 Q28XH2 A0A0N0U5T5
Ontologies
Topology
Subcellular location
Membrane
SignalP
Position: 1 - 25,
Likelihood: 0.956046
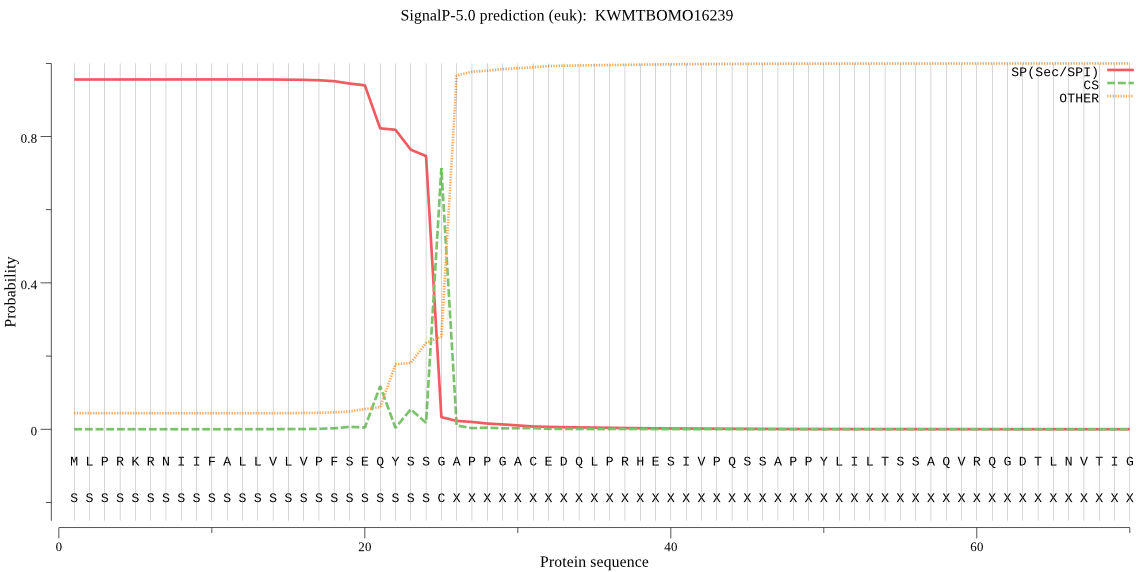
Length:
634
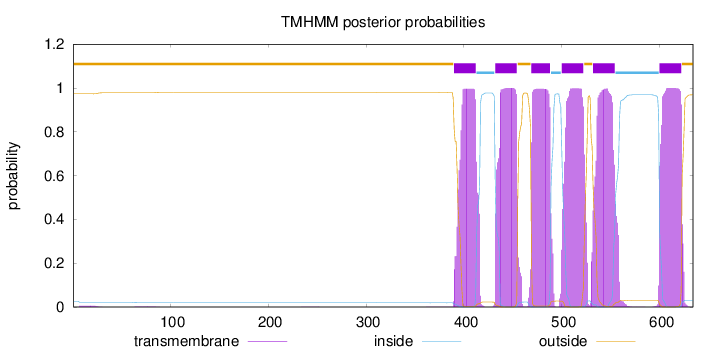
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
131.93439
Exp number, first 60 AAs:
0.09971
Total prob of N-in:
0.02652
outside
1 - 389
TMhelix
390 - 412
inside
413 - 431
TMhelix
432 - 454
outside
455 - 468
TMhelix
469 - 488
inside
489 - 499
TMhelix
500 - 522
outside
523 - 531
TMhelix
532 - 554
inside
555 - 599
TMhelix
600 - 622
outside
623 - 634
Population Genetic Test Statistics
Pi
194.711547
Theta
153.05357
Tajima's D
1.137598
CLR
0.0488
CSRT
0.699065046747663
Interpretation
Uncertain
