Gene
KWMTBOMO16238
Pre Gene Modal
BGIBMGA013987
Annotation
DNA_polymerase_epsilon_subunit_2_[Papilio_xuthus]
Full name
DNA polymerase epsilon subunit
+ More
DNA polymerase epsilon subunit 2
DNA polymerase epsilon subunit 2
Alternative Name
DNA polymerase II subunit 2
DNA polymerase epsilon subunit B
DNA polymerase epsilon subunit B
Location in the cell
Nuclear Reliability : 1.942
Sequence
CDS
ATGACTCCACATGAACGACGGGCAATTCTTGACAAGTTAACTTCACACATTTTAAAACAATGTATTTCACAGCCCGTATTAGAGAAAGAACACCTGGAAATCGCCTTCAAAGAATGCTCTGCATCGGGTCTTGAAGAGACCGAAACTGTTTTAAACGTAATCGACGCTTTTAACATTCCCAAACTGAGTTACGACAATGAGAGAAATAAGTTCGTTAAATCTGTACATAAGAACAATTTATATCCAGAACCGAAGTGGAAAGCGCAATTCTTGATAGACAGATACGAAATAATATGGCAAAGAACTGTTAGGAATAAATTGTTTGCTCAAGAAGCCTTGCCTTCATTGGCCGAGGAGAAATATTTCCAATTGCGCAAAATCGAAGCGCTGCTAAGCTCTTCTAGTCGAATCGATGATGTGATTGTGCTCGGTCTTTTAATACAGTTGACAGAAGGAAAATATTACTTGGAAGATCCAACTGGCAGTGTTGTTCTCGATATGTCACAAACAAGGTACCACTCAGGCTTATTCACTGAATATAGCTTCGTTTTAGTTGAAGGTTATTATGATGACAAGGTATTAAATGTTATGGGCCTTGTGCTACCGCCGCCGGAAAGCAGAGCCTCGTCACTGCCATTTTTTGGTAATTTAAATACATTTGGTGGAAATTCTAAGACTTTACTTAAAAACTCTAAGTCATTGCTAAAGATTGAACAGGAAAATAAAGAAGGAATGATTATATTCTTATCTGATGTTTGGATAGACACATTGAAAGTTATGAATAACTTAAGAACATTATTTAGTGGCTACAATGATTTTCCACCTGTGGCTATCGTGTTCATGGGTGAATTCCTCTCGTGTCCGTATGGTCAGGAGCATTGCACACAGCTACGAATGGCATTATCAAACTTAGGTGAAATAATAGCTCAATTCAAGAAATTGAAAGATGAATGCAAATTTATCTTTGTACCTGGCAGATCGGATCCTTGTGCTGCCAATGTTTTACCAAGACCAGCAATTCCAAGTTCCATAACACAAGGTATAAGAGAAAAATTAGGTGATGCAGTCATTTTCACAACGAACCCATGCAGAATACAATACTGTACACAAGAGATAGTAGTTTTCAGGCAAGATTTAGTAATGAAAATGTGTAGGAATTCTGTACATTTTCCGGAGACTGGTGATATTCCTGATCATTTGGTGAAGACTTTGTTTAGTCAATGTACACTGTCACCATTGTCATTAGCTATACAACCAGTGTATTGGAAGCATTCGGATTCTTTAAGTTTGTATCCGATGCCGGATTTGGTAGTGATTGGCGACCATTTCCAACCTTATACTCGCTCATATCAAAACTCGCAGATTATGAACCCTGGGTCTTTTCCGCGAACTGAGTTCTCATTTAAAGTGTATGTACCAAGTACGAAAACAGTTGAAGATTCACAAATACCGAAAGATGATGGATGA
Protein
MTPHERRAILDKLTSHILKQCISQPVLEKEHLEIAFKECSASGLEETETVLNVIDAFNIPKLSYDNERNKFVKSVHKNNLYPEPKWKAQFLIDRYEIIWQRTVRNKLFAQEALPSLAEEKYFQLRKIEALLSSSSRIDDVIVLGLLIQLTEGKYYLEDPTGSVVLDMSQTRYHSGLFTEYSFVLVEGYYDDKVLNVMGLVLPPPESRASSLPFFGNLNTFGGNSKTLLKNSKSLLKIEQENKEGMIIFLSDVWIDTLKVMNNLRTLFSGYNDFPPVAIVFMGEFLSCPYGQEHCTQLRMALSNLGEIIAQFKKLKDECKFIFVPGRSDPCAANVLPRPAIPSSITQGIREKLGDAVIFTTNPCRIQYCTQEIVVFRQDLVMKMCRNSVHFPETGDIPDHLVKTLFSQCTLSPLSLAIQPVYWKHSDSLSLYPMPDLVVIGDHFQPYTRSYQNSQIMNPGSFPRTEFSFKVYVPSTKTVEDSQIPKDDG
Summary
Description
Participates in DNA repair and in chromosomal DNA replication.
Catalytic Activity
a 2'-deoxyribonucleoside 5'-triphosphate + DNA(n) = diphosphate + DNA(n+1)
Subunit
Component of the epsilon DNA polymerase complex consisting of four subunits: POLE, POLE2, POLE3 and POLE4.
Consists of four subunits: POLE, POLE2, POLE3 and POLE4.
Consists of four subunits: POLE, POLE2, POLE3 and POLE4.
Miscellaneous
In eukaryotes there are five DNA polymerases: alpha, beta, gamma, delta, and epsilon which are responsible for different reactions of DNA synthesis.
Similarity
Belongs to the DNA polymerase epsilon subunit B family.
Keywords
3D-structure
Alternative splicing
Complete proteome
DNA replication
DNA-binding
DNA-directed DNA polymerase
Nucleotidyltransferase
Nucleus
Polymorphism
Reference proteome
Transferase
Feature
chain DNA polymerase epsilon subunit 2
splice variant In isoform 2.
sequence variant In dbSNP:rs34857719.
splice variant In isoform 2.
sequence variant In dbSNP:rs34857719.
Uniprot
H9JWS3
A0A0N1IN55
A0A0N1IFG7
A0A1E1WDM0
A0A3S2TG39
A0A212EKM9
+ More
A0A0L7LN72 A0A2A4JXT9 A0A2H1WB86 A0A212FF07 A0A067QG57 A0A2P8XZD3 A0A210PQ05 A0A2J7PDH2 V4A559 A0A2C9JIZ0 W4YG06 A0A1B6EGQ5 T1FST1 A0A1B6LQV0 A0A1B0GQJ6 A0A1B0CS94 A0A182NML0 A0A1B6M9S3 A0A1B6LUI4 A0A384AZP2 A0A182YET4 A0A0A9X6F9 I3M8Z7 A0A182Q0Z7 P56282 F1SHZ5 A0A2K5JNE0 Q9DGB4 E2RCT9 A0A336M4A5 A0A2K6B0N7 A0A2K5LZS5 A0A2Y9MVC1 A0A3Q7NWF1 G1RN11 G3RX47 A0A2U3V0L9 A0A2R9ATT1 A0A341A9T5 A0A0D9RRV3 G7MXP9 A0A2Y9EN08 A0A2C9JIX3 A0A1L8F9Q8 A0A2J7PDF8 G1NLS9 A0A2Y9I4B9 A0A2U3XDY1 J9PBE8 A0A2J8WKD0 A0A3B3QMI3 H0V2V0 Q5BJ62 A0A340WDB8 A0A336MRN2 A0A2K6P4M8 M3WAU8 A0A182VPQ8 A0A096NUT6 A0A3Q7T7M8 K7C803 W5N091 H9G4G4 A0A2K5S983 A0A2K5U0M8 A0A2K5U0N6 H2Q891 A0A1S3HPG0 A0A182PSQ3 Q3TA87 A0A2T7NYS2 M3Y0R0 F7ET97 A0A3Q7Y075 U3DJM7 A0A091DH04 W5QHE5 Q5ZKQ6 A0A2Y9I667 A0A2U3XDY2 A0A1Q3EVR8 A0A384CCH0 A0A2Y9IRC2 F1MKI8 A0A2K5CWV8 A0A3Q7NWI2 U3IIN1 S4RLY7 G9KHP1 A7YWS7 O54956 A0A2K6LRW4 F6ZV13 A0A182GFR6
A0A0L7LN72 A0A2A4JXT9 A0A2H1WB86 A0A212FF07 A0A067QG57 A0A2P8XZD3 A0A210PQ05 A0A2J7PDH2 V4A559 A0A2C9JIZ0 W4YG06 A0A1B6EGQ5 T1FST1 A0A1B6LQV0 A0A1B0GQJ6 A0A1B0CS94 A0A182NML0 A0A1B6M9S3 A0A1B6LUI4 A0A384AZP2 A0A182YET4 A0A0A9X6F9 I3M8Z7 A0A182Q0Z7 P56282 F1SHZ5 A0A2K5JNE0 Q9DGB4 E2RCT9 A0A336M4A5 A0A2K6B0N7 A0A2K5LZS5 A0A2Y9MVC1 A0A3Q7NWF1 G1RN11 G3RX47 A0A2U3V0L9 A0A2R9ATT1 A0A341A9T5 A0A0D9RRV3 G7MXP9 A0A2Y9EN08 A0A2C9JIX3 A0A1L8F9Q8 A0A2J7PDF8 G1NLS9 A0A2Y9I4B9 A0A2U3XDY1 J9PBE8 A0A2J8WKD0 A0A3B3QMI3 H0V2V0 Q5BJ62 A0A340WDB8 A0A336MRN2 A0A2K6P4M8 M3WAU8 A0A182VPQ8 A0A096NUT6 A0A3Q7T7M8 K7C803 W5N091 H9G4G4 A0A2K5S983 A0A2K5U0M8 A0A2K5U0N6 H2Q891 A0A1S3HPG0 A0A182PSQ3 Q3TA87 A0A2T7NYS2 M3Y0R0 F7ET97 A0A3Q7Y075 U3DJM7 A0A091DH04 W5QHE5 Q5ZKQ6 A0A2Y9I667 A0A2U3XDY2 A0A1Q3EVR8 A0A384CCH0 A0A2Y9IRC2 F1MKI8 A0A2K5CWV8 A0A3Q7NWI2 U3IIN1 S4RLY7 G9KHP1 A7YWS7 O54956 A0A2K6LRW4 F6ZV13 A0A182GFR6
EC Number
2.7.7.7
Pubmed
19121390
26354079
22118469
26227816
24845553
29403074
+ More
28812685 23254933 15562597 25244985 25401762 26823975 9405441 9443964 11433027 14702039 12508121 15489334 10801849 30723633 11296256 16341006 22398555 22722832 22002653 27762356 20838655 29240929 21993624 25362486 17975172 21881562 16136131 10349636 11042159 11076861 11217851 12466851 16141073 25243066 20809919 15642098 19393038 23749191 23236062 16141072 17431167 26483478
28812685 23254933 15562597 25244985 25401762 26823975 9405441 9443964 11433027 14702039 12508121 15489334 10801849 30723633 11296256 16341006 22398555 22722832 22002653 27762356 20838655 29240929 21993624 25362486 17975172 21881562 16136131 10349636 11042159 11076861 11217851 12466851 16141073 25243066 20809919 15642098 19393038 23749191 23236062 16141072 17431167 26483478
EMBL
BABH01018560
KQ459166
KPJ03544.1
KQ460689
KPJ12746.1
GDQN01005974
+ More
JAT85080.1 RSAL01000177 RVE45027.1 AGBW02014237 OWR42041.1 JTDY01000478 KOB76998.1 NWSH01000468 PCG76202.1 ODYU01007498 SOQ50320.1 AGBW02008869 OWR52329.1 KK853567 KDR06663.1 PYGN01001128 PSN37360.1 NEDP02005562 OWF38563.1 NEVH01026387 PNF14373.1 KB202481 ESO90150.1 AAGJ04112717 GEDC01019285 GEDC01000244 JAS18013.1 JAS37054.1 AMQM01003729 KB096275 ESO06951.1 GEBQ01013907 JAT26070.1 AJVK01017154 AJWK01025807 GEBQ01007306 JAT32671.1 GEBQ01012655 JAT27322.1 GBHO01029196 GBHO01020976 GDHC01007572 JAG14408.1 JAG22628.1 JAQ11057.1 AGTP01067338 AGTP01067339 AGTP01067340 AGTP01067341 AGTP01067342 AXCN02001597 AF025840 AF036899 AF387034 AF387021 AF387022 AF387023 AF387024 AF387025 AF387026 AF387027 AF387028 AF387029 AF387030 AF387031 AF387032 AF387033 EF506887 AK293163 AL139099 AL591767 BC112962 BC126218 BC126220 AEMK02000004 DQIR01027653 DQIR01205437 DQIR01312570 HCZ83128.1 BC097773 AB048257 AAH97773.1 BAB12726.1 AAEX03005717 AAEX03005718 UFQT01000542 SSX25106.1 ADFV01039976 ADFV01039977 ADFV01039978 ADFV01039979 ADFV01039980 CABD030092223 CABD030092224 CABD030092225 CABD030092226 CABD030092227 CABD030092228 CABD030092229 CABD030092230 AJFE02112244 AJFE02112245 AJFE02112246 AJFE02112247 AJFE02112248 AJFE02112249 AQIB01058976 AQIB01058977 AQIB01058978 AQIB01058979 AQIB01058980 AQIB01058981 AQIB01058982 AQIB01058983 AQIB01058984 CM001259 EHH27857.1 CM004480 OCT68329.1 PNF14374.1 NDHI03003386 PNJ70225.1 AAKN02027602 AAKN02027603 AAKN02027604 BC091608 AAH91608.1 UFQS01001948 UFQT01001948 SSX12769.1 SSX32211.1 AANG04004459 AHZZ02032139 AHZZ02032140 AHZZ02032141 GABF01007108 JAA15037.1 AHAT01003708 AAWZ02030255 AAWZ02030256 AQIA01064818 AQIA01064819 AQIA01064820 AC191868 NBAG03000214 PNI83282.1 AK172024 BAE42782.1 PZQS01000008 PVD26327.1 AEYP01003513 AEYP01003514 AEYP01003515 AEYP01003516 GAMP01005684 JAB47071.1 GAMS01009545 JAB13591.1 KN122569 KFO29738.1 AMGL01102946 AJ720028 GFDL01015653 JAV19392.1 ADON01057939 JP015818 AES04416.1 BC134762 AK077592 BC063772 AF036898 JSUE03039694 JXUM01060178 JXUM01060179 KQ562091 KXJ76713.1
JAT85080.1 RSAL01000177 RVE45027.1 AGBW02014237 OWR42041.1 JTDY01000478 KOB76998.1 NWSH01000468 PCG76202.1 ODYU01007498 SOQ50320.1 AGBW02008869 OWR52329.1 KK853567 KDR06663.1 PYGN01001128 PSN37360.1 NEDP02005562 OWF38563.1 NEVH01026387 PNF14373.1 KB202481 ESO90150.1 AAGJ04112717 GEDC01019285 GEDC01000244 JAS18013.1 JAS37054.1 AMQM01003729 KB096275 ESO06951.1 GEBQ01013907 JAT26070.1 AJVK01017154 AJWK01025807 GEBQ01007306 JAT32671.1 GEBQ01012655 JAT27322.1 GBHO01029196 GBHO01020976 GDHC01007572 JAG14408.1 JAG22628.1 JAQ11057.1 AGTP01067338 AGTP01067339 AGTP01067340 AGTP01067341 AGTP01067342 AXCN02001597 AF025840 AF036899 AF387034 AF387021 AF387022 AF387023 AF387024 AF387025 AF387026 AF387027 AF387028 AF387029 AF387030 AF387031 AF387032 AF387033 EF506887 AK293163 AL139099 AL591767 BC112962 BC126218 BC126220 AEMK02000004 DQIR01027653 DQIR01205437 DQIR01312570 HCZ83128.1 BC097773 AB048257 AAH97773.1 BAB12726.1 AAEX03005717 AAEX03005718 UFQT01000542 SSX25106.1 ADFV01039976 ADFV01039977 ADFV01039978 ADFV01039979 ADFV01039980 CABD030092223 CABD030092224 CABD030092225 CABD030092226 CABD030092227 CABD030092228 CABD030092229 CABD030092230 AJFE02112244 AJFE02112245 AJFE02112246 AJFE02112247 AJFE02112248 AJFE02112249 AQIB01058976 AQIB01058977 AQIB01058978 AQIB01058979 AQIB01058980 AQIB01058981 AQIB01058982 AQIB01058983 AQIB01058984 CM001259 EHH27857.1 CM004480 OCT68329.1 PNF14374.1 NDHI03003386 PNJ70225.1 AAKN02027602 AAKN02027603 AAKN02027604 BC091608 AAH91608.1 UFQS01001948 UFQT01001948 SSX12769.1 SSX32211.1 AANG04004459 AHZZ02032139 AHZZ02032140 AHZZ02032141 GABF01007108 JAA15037.1 AHAT01003708 AAWZ02030255 AAWZ02030256 AQIA01064818 AQIA01064819 AQIA01064820 AC191868 NBAG03000214 PNI83282.1 AK172024 BAE42782.1 PZQS01000008 PVD26327.1 AEYP01003513 AEYP01003514 AEYP01003515 AEYP01003516 GAMP01005684 JAB47071.1 GAMS01009545 JAB13591.1 KN122569 KFO29738.1 AMGL01102946 AJ720028 GFDL01015653 JAV19392.1 ADON01057939 JP015818 AES04416.1 BC134762 AK077592 BC063772 AF036898 JSUE03039694 JXUM01060178 JXUM01060179 KQ562091 KXJ76713.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000283053
UP000007151
UP000037510
+ More
UP000218220 UP000027135 UP000245037 UP000242188 UP000235965 UP000030746 UP000076420 UP000007110 UP000015101 UP000092462 UP000092461 UP000075884 UP000261681 UP000076408 UP000005215 UP000075886 UP000005640 UP000008227 UP000233080 UP000002254 UP000233120 UP000233060 UP000248483 UP000286641 UP000001073 UP000001519 UP000245320 UP000240080 UP000252040 UP000029965 UP000248484 UP000186698 UP000001645 UP000248481 UP000245341 UP000261540 UP000005447 UP000265300 UP000233200 UP000011712 UP000075920 UP000028761 UP000286640 UP000018468 UP000001646 UP000233040 UP000233100 UP000002277 UP000085678 UP000075885 UP000245119 UP000000715 UP000008225 UP000286642 UP000028990 UP000002356 UP000000539 UP000261680 UP000248482 UP000009136 UP000233020 UP000016666 UP000245300 UP000000589 UP000233180 UP000006718 UP000069940 UP000249989
UP000218220 UP000027135 UP000245037 UP000242188 UP000235965 UP000030746 UP000076420 UP000007110 UP000015101 UP000092462 UP000092461 UP000075884 UP000261681 UP000076408 UP000005215 UP000075886 UP000005640 UP000008227 UP000233080 UP000002254 UP000233120 UP000233060 UP000248483 UP000286641 UP000001073 UP000001519 UP000245320 UP000240080 UP000252040 UP000029965 UP000248484 UP000186698 UP000001645 UP000248481 UP000245341 UP000261540 UP000005447 UP000265300 UP000233200 UP000011712 UP000075920 UP000028761 UP000286640 UP000018468 UP000001646 UP000233040 UP000233100 UP000002277 UP000085678 UP000075885 UP000245119 UP000000715 UP000008225 UP000286642 UP000028990 UP000002356 UP000000539 UP000261680 UP000248482 UP000009136 UP000233020 UP000016666 UP000245300 UP000000589 UP000233180 UP000006718 UP000069940 UP000249989
Interpro
SUPFAM
SSF101756
SSF101756
ProteinModelPortal
H9JWS3
A0A0N1IN55
A0A0N1IFG7
A0A1E1WDM0
A0A3S2TG39
A0A212EKM9
+ More
A0A0L7LN72 A0A2A4JXT9 A0A2H1WB86 A0A212FF07 A0A067QG57 A0A2P8XZD3 A0A210PQ05 A0A2J7PDH2 V4A559 A0A2C9JIZ0 W4YG06 A0A1B6EGQ5 T1FST1 A0A1B6LQV0 A0A1B0GQJ6 A0A1B0CS94 A0A182NML0 A0A1B6M9S3 A0A1B6LUI4 A0A384AZP2 A0A182YET4 A0A0A9X6F9 I3M8Z7 A0A182Q0Z7 P56282 F1SHZ5 A0A2K5JNE0 Q9DGB4 E2RCT9 A0A336M4A5 A0A2K6B0N7 A0A2K5LZS5 A0A2Y9MVC1 A0A3Q7NWF1 G1RN11 G3RX47 A0A2U3V0L9 A0A2R9ATT1 A0A341A9T5 A0A0D9RRV3 G7MXP9 A0A2Y9EN08 A0A2C9JIX3 A0A1L8F9Q8 A0A2J7PDF8 G1NLS9 A0A2Y9I4B9 A0A2U3XDY1 J9PBE8 A0A2J8WKD0 A0A3B3QMI3 H0V2V0 Q5BJ62 A0A340WDB8 A0A336MRN2 A0A2K6P4M8 M3WAU8 A0A182VPQ8 A0A096NUT6 A0A3Q7T7M8 K7C803 W5N091 H9G4G4 A0A2K5S983 A0A2K5U0M8 A0A2K5U0N6 H2Q891 A0A1S3HPG0 A0A182PSQ3 Q3TA87 A0A2T7NYS2 M3Y0R0 F7ET97 A0A3Q7Y075 U3DJM7 A0A091DH04 W5QHE5 Q5ZKQ6 A0A2Y9I667 A0A2U3XDY2 A0A1Q3EVR8 A0A384CCH0 A0A2Y9IRC2 F1MKI8 A0A2K5CWV8 A0A3Q7NWI2 U3IIN1 S4RLY7 G9KHP1 A7YWS7 O54956 A0A2K6LRW4 F6ZV13 A0A182GFR6
A0A0L7LN72 A0A2A4JXT9 A0A2H1WB86 A0A212FF07 A0A067QG57 A0A2P8XZD3 A0A210PQ05 A0A2J7PDH2 V4A559 A0A2C9JIZ0 W4YG06 A0A1B6EGQ5 T1FST1 A0A1B6LQV0 A0A1B0GQJ6 A0A1B0CS94 A0A182NML0 A0A1B6M9S3 A0A1B6LUI4 A0A384AZP2 A0A182YET4 A0A0A9X6F9 I3M8Z7 A0A182Q0Z7 P56282 F1SHZ5 A0A2K5JNE0 Q9DGB4 E2RCT9 A0A336M4A5 A0A2K6B0N7 A0A2K5LZS5 A0A2Y9MVC1 A0A3Q7NWF1 G1RN11 G3RX47 A0A2U3V0L9 A0A2R9ATT1 A0A341A9T5 A0A0D9RRV3 G7MXP9 A0A2Y9EN08 A0A2C9JIX3 A0A1L8F9Q8 A0A2J7PDF8 G1NLS9 A0A2Y9I4B9 A0A2U3XDY1 J9PBE8 A0A2J8WKD0 A0A3B3QMI3 H0V2V0 Q5BJ62 A0A340WDB8 A0A336MRN2 A0A2K6P4M8 M3WAU8 A0A182VPQ8 A0A096NUT6 A0A3Q7T7M8 K7C803 W5N091 H9G4G4 A0A2K5S983 A0A2K5U0M8 A0A2K5U0N6 H2Q891 A0A1S3HPG0 A0A182PSQ3 Q3TA87 A0A2T7NYS2 M3Y0R0 F7ET97 A0A3Q7Y075 U3DJM7 A0A091DH04 W5QHE5 Q5ZKQ6 A0A2Y9I667 A0A2U3XDY2 A0A1Q3EVR8 A0A384CCH0 A0A2Y9IRC2 F1MKI8 A0A2K5CWV8 A0A3Q7NWI2 U3IIN1 S4RLY7 G9KHP1 A7YWS7 O54956 A0A2K6LRW4 F6ZV13 A0A182GFR6
PDB
5VBN
E-value=1.53604e-123,
Score=1134
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Nucleus
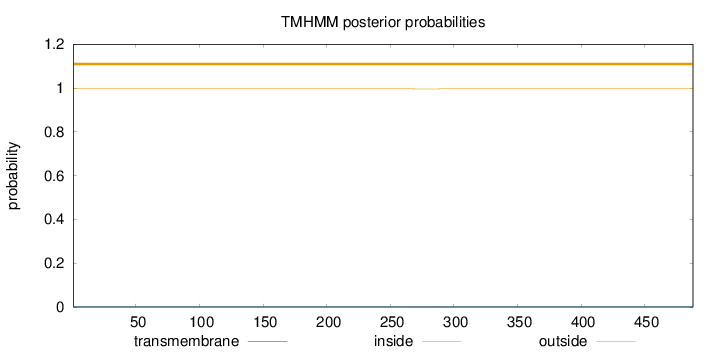
Length:
488
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01566
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00375
outside
1 - 488
Population Genetic Test Statistics
Pi
190.495258
Theta
179.77271
Tajima's D
0.187659
CLR
0.219374
CSRT
0.425778711064447
Interpretation
Uncertain
