Gene
KWMTBOMO16237
Pre Gene Modal
BGIBMGA013909
Annotation
PREDICTED:_uncharacterized_protein_LOC101735359_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.523 Mitochondrial Reliability : 1.323 Nuclear Reliability : 1.304
Sequence
CDS
ATGGATTTAGCAAAATTATTAGAAGATCTTCCTTGGATAAAACTTGACTGGTGTTTTCCGAGCATGACCCAAGAGACCAATGAACTTATTCAGAAATGCACTGAAATACCTGAAATTGAACCAGACCCTGAAGTTGATGAAATTATTGAGAGAAGCAAGAATTTTCCTATACCTTTCCCGATTCAGACTGTGAGATTGGAGAAACTGAAGGAGCATCGTCCCATAGACAGATTAAAGAGGAATATTTCAGAGACATATCCTATTATACACGAGAGGGTCTTGTTGCTGATGGCACACTTTTTAATATATAAACGTGAACATGGCAGTAGTATAGAGAAAGAATTATACAGAGACATGACCATACCAGAGCTTATTGATCGATTATTACTGAAAAGAGCCGTATCGTTTATGGGAGCTCGCGATGCATATATGTTAATGAGTGGCAAGAAGGGCGTCGATGGTTGGGAGAACGTGGGGACCCCGGCGGAAACAGAGCCGCTGGTGTTGAAAGACGTGCTCAGCTACGATGAGATAAAGCTCTCTGCTTTTTTGTTCGTGAGCGGACCCACCGAGTGCATCAACAGTGGCTCCAGGAGGAACTGCGGCGTTCTAGACGACGATGACATCGAGAAGGAAGCCATTATCATAGGAGCCATAGGGCCGCGCTTCAAGCGGTTGAATCGAATGGACTACGAGGACATGGTGATATCGAAAACGCAGAACACTGCAGAGCGCGGCTACGGCGAGCACGAGGCGCCGACCAGGTGCATGGACGTACTGCGACACGCGTACACGCGCGACGCCAGCCTCGCCAAGCGCGCCTGGAGGCAGCTGTGGGCCGAGCTGTACCAGGTGCACAGCTACACGTACGAGGAGCTGAGCGCGCGGCTCGCGGGGGCGGCCTCGGACCGCTACGTGAAGCTGCCGCGGGGGGGCGCCTGGTTCGACAACGAGGTCTACTACAAGCGGATCTGCATCCTCGCGGAGACCGTGCTGCTGGAGGCCGAGGGGCGGGCTCGAGGCCGGAGCGTGTTCCTCAACGTGGTGGGCTGCGGACTGGGAGTGTGGAAGATATCCCCTCACCAGACCGACGTGTACGTGCTCACCTTCCTCGAACGCATCCGCGCCATGCTGGACGAGGAGGCACTCGATCACATCACAGACGTCAACTTCGCCTATATTGGTACTTCAAAGAGTGTTACAGCTTTATTCGCGGATCGAAGTGAAGACAAAGAGAGTGCCGCTAAGATAATGTTTCTGAAGAACGAGAGACATCCGAAGGGTGGCGTGAGCGTGCAGCTGGAGGACAGGGAGCCGGCGAGCAGGCTGCACGGGCGGCACCGGGGCAAGCTGCTCGTCCTCACCTATCCCTGGGACGGGAACGCGCAGCCGGGGAACGAGTTCTGGGAGGAAAGTTTGTCGAGTTCAGGCGACCCGGCGGCCGCGTGCTCCACGCAAGTGTCGGAATTGCACAACGCGCGCGGTGCGGGTAGCCGCACGCGGCGGCGTGCACGCGCTCCGGGACCGCGCGCGGCCGCTCACACCTGTGTCGCTTAG
Protein
MDLAKLLEDLPWIKLDWCFPSMTQETNELIQKCTEIPEIEPDPEVDEIIERSKNFPIPFPIQTVRLEKLKEHRPIDRLKRNISETYPIIHERVLLLMAHFLIYKREHGSSIEKELYRDMTIPELIDRLLLKRAVSFMGARDAYMLMSGKKGVDGWENVGTPAETEPLVLKDVLSYDEIKLSAFLFVSGPTECINSGSRRNCGVLDDDDIEKEAIIIGAIGPRFKRLNRMDYEDMVISKTQNTAERGYGEHEAPTRCMDVLRHAYTRDASLAKRAWRQLWAELYQVHSYTYEELSARLAGAASDRYVKLPRGGAWFDNEVYYKRICILAETVLLEAEGRARGRSVFLNVVGCGLGVWKISPHQTDVYVLTFLERIRAMLDEEALDHITDVNFAYIGTSKSVTALFADRSEDKESAAKIMFLKNERHPKGGVSVQLEDREPASRLHGRHRGKLLVLTYPWDGNAQPGNEFWEESLSSSGDPAAACSTQVSELHNARGAGSRTRRRARAPGPRAAAHTCVA
Summary
Uniprot
H9JWJ5
H9JWS2
A0A0N1PF66
A0A0N1I8S1
A0A2H1WB77
A0A2W1BTJ0
+ More
A0A212EKN2 S4PGH9 A0A1Y1MEH5 A0A1Y1MAX2 A0A0L7LVN1 A0A0T6B964 A0A1Y1KTF3 A0A034VHV9 A0A034VII7 A0A034VR60 A0A2J7QND3 A0A0A1WMZ7 D2A3W8 A0A2J7Q8F1 A0A0A1X237 A0A0A1XQ92 A0A0A1XRY2 A0A0M9A2F5 E9J8R2 A0A1Y1MW12 W8BBD8 A0A310S6I1 W8BF98 A0A087ZQ89 A0A2P8Y6C6 E1ZX59 A0A0L7R433 A0A154PH72 A0A1I8NIN0 A0A0K8V8T2 E2BNU5 F4X5P0 A0A034WC84 A0A1W4V1T8 A0A0K8V2Z8 A0A1B0ATB3 A0A1A9XAJ7 W8BSI2 N6UAA7 T1P871 D6WR72 B4JKR6 A0A1A9X4N1 N6T6A5 A0A1I8MRE2 B4L2F9 A0A1I8PU58 B4JKR7 A0A067QZ28 A0A2P8Y6D4 A0A0K8U4F6 A0A1I8P191 A0A0C9R8Z9 A0A1L8ECA9 B4R4I5 A0A1L8EC02 A0A1L8ECC1 A0A1I8NEK6 A0A0K8VLN8 A0A034VGX6 Q9W4J9 B3NU36 B4MKU4 A0A195BYZ9 W8AUL8 B4NPT8 B4H2H6 Q29IH6 A0A3B0K4S3 A0A034VGX1 A0A0L0BNK8 J3JUT6 B4MEE0 A0A151K1R1 B3MXL8 W8AIK8 A0A1I8NIL9 A0A1W4V7F3 B4I0Y6 A0A0M5J3J0 A0A0A1WWH6 A0A151J2C4 A0A1I8P182 A0A151IHB0 F4WAU2 B4MED9 A0A1I8P145 A0A3B0JXH6 B3MXX2 A0A0Q9WG97 B4L2F8 B4Q051
A0A212EKN2 S4PGH9 A0A1Y1MEH5 A0A1Y1MAX2 A0A0L7LVN1 A0A0T6B964 A0A1Y1KTF3 A0A034VHV9 A0A034VII7 A0A034VR60 A0A2J7QND3 A0A0A1WMZ7 D2A3W8 A0A2J7Q8F1 A0A0A1X237 A0A0A1XQ92 A0A0A1XRY2 A0A0M9A2F5 E9J8R2 A0A1Y1MW12 W8BBD8 A0A310S6I1 W8BF98 A0A087ZQ89 A0A2P8Y6C6 E1ZX59 A0A0L7R433 A0A154PH72 A0A1I8NIN0 A0A0K8V8T2 E2BNU5 F4X5P0 A0A034WC84 A0A1W4V1T8 A0A0K8V2Z8 A0A1B0ATB3 A0A1A9XAJ7 W8BSI2 N6UAA7 T1P871 D6WR72 B4JKR6 A0A1A9X4N1 N6T6A5 A0A1I8MRE2 B4L2F9 A0A1I8PU58 B4JKR7 A0A067QZ28 A0A2P8Y6D4 A0A0K8U4F6 A0A1I8P191 A0A0C9R8Z9 A0A1L8ECA9 B4R4I5 A0A1L8EC02 A0A1L8ECC1 A0A1I8NEK6 A0A0K8VLN8 A0A034VGX6 Q9W4J9 B3NU36 B4MKU4 A0A195BYZ9 W8AUL8 B4NPT8 B4H2H6 Q29IH6 A0A3B0K4S3 A0A034VGX1 A0A0L0BNK8 J3JUT6 B4MEE0 A0A151K1R1 B3MXL8 W8AIK8 A0A1I8NIL9 A0A1W4V7F3 B4I0Y6 A0A0M5J3J0 A0A0A1WWH6 A0A151J2C4 A0A1I8P182 A0A151IHB0 F4WAU2 B4MED9 A0A1I8P145 A0A3B0JXH6 B3MXX2 A0A0Q9WG97 B4L2F8 B4Q051
Pubmed
19121390
26354079
28756777
22118469
23622113
28004739
+ More
26227816 25348373 25830018 18362917 19820115 21282665 24495485 29403074 20798317 25315136 21719571 23537049 17994087 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 26108605 22516182 18057021 17550304
26227816 25348373 25830018 18362917 19820115 21282665 24495485 29403074 20798317 25315136 21719571 23537049 17994087 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 26108605 22516182 18057021 17550304
EMBL
BABH01018558
BABH01018559
BABH01018560
BABH01018557
KQ459166
KPJ03543.1
+ More
KQ460689 KPJ12747.1 ODYU01007498 SOQ50318.1 KZ149970 PZC76090.1 AGBW02014237 OWR42043.1 GAIX01003657 JAA88903.1 GEZM01036097 JAV82980.1 GEZM01036098 JAV82979.1 JTDY01000039 KOB79241.1 LJIG01009049 KRT83858.1 GEZM01074086 JAV64679.1 GAKP01017593 GAKP01017592 JAC41360.1 GAKP01017589 JAC41363.1 GAKP01013186 JAC45766.1 NEVH01013195 PNF30104.1 GBXI01014231 JAD00061.1 KQ971348 EFA04874.1 NEVH01016959 PNF24846.1 GBXI01008898 JAD05394.1 GBXI01001584 JAD12708.1 GBXI01000555 JAD13737.1 KQ435756 KOX75900.1 GL769036 EFZ10832.1 GEZM01019121 JAV89864.1 GAMC01010573 JAB95982.1 KQ767768 OAD53304.1 GAMC01018161 JAB88394.1 PYGN01000874 PSN39812.1 GL435030 EFN74149.1 KQ414658 KOC65650.1 KQ434904 KZC11193.1 GDHF01028203 GDHF01017058 JAI24111.1 JAI35256.1 GL449507 EFN82628.1 GL888730 EGI58245.1 GAKP01007237 JAC51715.1 GDHF01019032 JAI33282.1 JXJN01003232 GAMC01006687 GAMC01006686 JAB99868.1 APGK01032677 KB740848 ENN78655.1 KA644774 AFP59403.1 KQ971354 EFA06000.1 CH916370 EDW00169.1 APGK01042182 APGK01042183 KB741002 ENN75744.1 CH933810 EDW06835.2 EDW00170.1 KK852815 KDR15804.1 PSN39813.1 GDHF01031119 JAI21195.1 GBYB01012829 JAG82596.1 GFDG01002533 JAV16266.1 CM000366 EDX17075.1 GFDG01002532 JAV16267.1 GFDG01002534 JAV16265.1 GDHF01012517 JAI39797.1 GAKP01017590 JAC41362.1 AE014298 AY113290 AAF45952.2 AAM29295.1 CH954180 EDV45812.1 CH963847 EDW72800.1 KQ976394 KYM93143.1 GAMC01018162 JAB88393.1 CH964291 EDW86528.1 CH479204 EDW30543.1 CH379063 EAL32677.2 OUUW01000003 SPP78468.1 GAKP01017595 JAC41357.1 JRES01001596 KNC21627.1 BT127001 AEE61963.1 CH940664 EDW62915.1 KQ981225 KYN44376.1 CH902630 EDV38483.1 GAMC01018160 JAB88395.1 CH480819 EDW53167.1 CP012528 ALC49636.1 GBXI01011080 JAD03212.1 KQ980404 KYN16193.1 KQ977636 KYN01149.1 GL888053 EGI68698.1 EDW62914.2 KRF80850.1 KRF80851.1 SPP78469.1 EDV38587.1 KRF80849.1 EDW06834.1 CM000162 EDX01203.2
KQ460689 KPJ12747.1 ODYU01007498 SOQ50318.1 KZ149970 PZC76090.1 AGBW02014237 OWR42043.1 GAIX01003657 JAA88903.1 GEZM01036097 JAV82980.1 GEZM01036098 JAV82979.1 JTDY01000039 KOB79241.1 LJIG01009049 KRT83858.1 GEZM01074086 JAV64679.1 GAKP01017593 GAKP01017592 JAC41360.1 GAKP01017589 JAC41363.1 GAKP01013186 JAC45766.1 NEVH01013195 PNF30104.1 GBXI01014231 JAD00061.1 KQ971348 EFA04874.1 NEVH01016959 PNF24846.1 GBXI01008898 JAD05394.1 GBXI01001584 JAD12708.1 GBXI01000555 JAD13737.1 KQ435756 KOX75900.1 GL769036 EFZ10832.1 GEZM01019121 JAV89864.1 GAMC01010573 JAB95982.1 KQ767768 OAD53304.1 GAMC01018161 JAB88394.1 PYGN01000874 PSN39812.1 GL435030 EFN74149.1 KQ414658 KOC65650.1 KQ434904 KZC11193.1 GDHF01028203 GDHF01017058 JAI24111.1 JAI35256.1 GL449507 EFN82628.1 GL888730 EGI58245.1 GAKP01007237 JAC51715.1 GDHF01019032 JAI33282.1 JXJN01003232 GAMC01006687 GAMC01006686 JAB99868.1 APGK01032677 KB740848 ENN78655.1 KA644774 AFP59403.1 KQ971354 EFA06000.1 CH916370 EDW00169.1 APGK01042182 APGK01042183 KB741002 ENN75744.1 CH933810 EDW06835.2 EDW00170.1 KK852815 KDR15804.1 PSN39813.1 GDHF01031119 JAI21195.1 GBYB01012829 JAG82596.1 GFDG01002533 JAV16266.1 CM000366 EDX17075.1 GFDG01002532 JAV16267.1 GFDG01002534 JAV16265.1 GDHF01012517 JAI39797.1 GAKP01017590 JAC41362.1 AE014298 AY113290 AAF45952.2 AAM29295.1 CH954180 EDV45812.1 CH963847 EDW72800.1 KQ976394 KYM93143.1 GAMC01018162 JAB88393.1 CH964291 EDW86528.1 CH479204 EDW30543.1 CH379063 EAL32677.2 OUUW01000003 SPP78468.1 GAKP01017595 JAC41357.1 JRES01001596 KNC21627.1 BT127001 AEE61963.1 CH940664 EDW62915.1 KQ981225 KYN44376.1 CH902630 EDV38483.1 GAMC01018160 JAB88395.1 CH480819 EDW53167.1 CP012528 ALC49636.1 GBXI01011080 JAD03212.1 KQ980404 KYN16193.1 KQ977636 KYN01149.1 GL888053 EGI68698.1 EDW62914.2 KRF80850.1 KRF80851.1 SPP78469.1 EDV38587.1 KRF80849.1 EDW06834.1 CM000162 EDX01203.2
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000037510
UP000235965
+ More
UP000007266 UP000053105 UP000005203 UP000245037 UP000000311 UP000053825 UP000076502 UP000095301 UP000008237 UP000007755 UP000192221 UP000092460 UP000092443 UP000019118 UP000001070 UP000091820 UP000009192 UP000095300 UP000027135 UP000000304 UP000000803 UP000008711 UP000007798 UP000078540 UP000008744 UP000001819 UP000268350 UP000037069 UP000008792 UP000078541 UP000007801 UP000001292 UP000092553 UP000078492 UP000078542 UP000002282
UP000007266 UP000053105 UP000005203 UP000245037 UP000000311 UP000053825 UP000076502 UP000095301 UP000008237 UP000007755 UP000192221 UP000092460 UP000092443 UP000019118 UP000001070 UP000091820 UP000009192 UP000095300 UP000027135 UP000000304 UP000000803 UP000008711 UP000007798 UP000078540 UP000008744 UP000001819 UP000268350 UP000037069 UP000008792 UP000078541 UP000007801 UP000001292 UP000092553 UP000078492 UP000078542 UP000002282
Interpro
Gene 3D
ProteinModelPortal
H9JWJ5
H9JWS2
A0A0N1PF66
A0A0N1I8S1
A0A2H1WB77
A0A2W1BTJ0
+ More
A0A212EKN2 S4PGH9 A0A1Y1MEH5 A0A1Y1MAX2 A0A0L7LVN1 A0A0T6B964 A0A1Y1KTF3 A0A034VHV9 A0A034VII7 A0A034VR60 A0A2J7QND3 A0A0A1WMZ7 D2A3W8 A0A2J7Q8F1 A0A0A1X237 A0A0A1XQ92 A0A0A1XRY2 A0A0M9A2F5 E9J8R2 A0A1Y1MW12 W8BBD8 A0A310S6I1 W8BF98 A0A087ZQ89 A0A2P8Y6C6 E1ZX59 A0A0L7R433 A0A154PH72 A0A1I8NIN0 A0A0K8V8T2 E2BNU5 F4X5P0 A0A034WC84 A0A1W4V1T8 A0A0K8V2Z8 A0A1B0ATB3 A0A1A9XAJ7 W8BSI2 N6UAA7 T1P871 D6WR72 B4JKR6 A0A1A9X4N1 N6T6A5 A0A1I8MRE2 B4L2F9 A0A1I8PU58 B4JKR7 A0A067QZ28 A0A2P8Y6D4 A0A0K8U4F6 A0A1I8P191 A0A0C9R8Z9 A0A1L8ECA9 B4R4I5 A0A1L8EC02 A0A1L8ECC1 A0A1I8NEK6 A0A0K8VLN8 A0A034VGX6 Q9W4J9 B3NU36 B4MKU4 A0A195BYZ9 W8AUL8 B4NPT8 B4H2H6 Q29IH6 A0A3B0K4S3 A0A034VGX1 A0A0L0BNK8 J3JUT6 B4MEE0 A0A151K1R1 B3MXL8 W8AIK8 A0A1I8NIL9 A0A1W4V7F3 B4I0Y6 A0A0M5J3J0 A0A0A1WWH6 A0A151J2C4 A0A1I8P182 A0A151IHB0 F4WAU2 B4MED9 A0A1I8P145 A0A3B0JXH6 B3MXX2 A0A0Q9WG97 B4L2F8 B4Q051
A0A212EKN2 S4PGH9 A0A1Y1MEH5 A0A1Y1MAX2 A0A0L7LVN1 A0A0T6B964 A0A1Y1KTF3 A0A034VHV9 A0A034VII7 A0A034VR60 A0A2J7QND3 A0A0A1WMZ7 D2A3W8 A0A2J7Q8F1 A0A0A1X237 A0A0A1XQ92 A0A0A1XRY2 A0A0M9A2F5 E9J8R2 A0A1Y1MW12 W8BBD8 A0A310S6I1 W8BF98 A0A087ZQ89 A0A2P8Y6C6 E1ZX59 A0A0L7R433 A0A154PH72 A0A1I8NIN0 A0A0K8V8T2 E2BNU5 F4X5P0 A0A034WC84 A0A1W4V1T8 A0A0K8V2Z8 A0A1B0ATB3 A0A1A9XAJ7 W8BSI2 N6UAA7 T1P871 D6WR72 B4JKR6 A0A1A9X4N1 N6T6A5 A0A1I8MRE2 B4L2F9 A0A1I8PU58 B4JKR7 A0A067QZ28 A0A2P8Y6D4 A0A0K8U4F6 A0A1I8P191 A0A0C9R8Z9 A0A1L8ECA9 B4R4I5 A0A1L8EC02 A0A1L8ECC1 A0A1I8NEK6 A0A0K8VLN8 A0A034VGX6 Q9W4J9 B3NU36 B4MKU4 A0A195BYZ9 W8AUL8 B4NPT8 B4H2H6 Q29IH6 A0A3B0K4S3 A0A034VGX1 A0A0L0BNK8 J3JUT6 B4MEE0 A0A151K1R1 B3MXL8 W8AIK8 A0A1I8NIL9 A0A1W4V7F3 B4I0Y6 A0A0M5J3J0 A0A0A1WWH6 A0A151J2C4 A0A1I8P182 A0A151IHB0 F4WAU2 B4MED9 A0A1I8P145 A0A3B0JXH6 B3MXX2 A0A0Q9WG97 B4L2F8 B4Q051
Ontologies
PANTHER
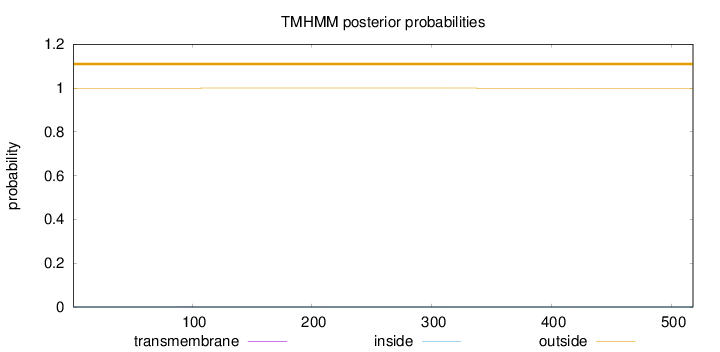
Topology
Length:
518
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00450999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00049
outside
1 - 518
Population Genetic Test Statistics
Pi
255.965617
Theta
19.057641
Tajima's D
-1.329266
CLR
251.229637
CSRT
0.0831458427078646
Interpretation
Uncertain
