Gene
KWMTBOMO16235
Pre Gene Modal
BGIBMGA013910
Annotation
PREDICTED:_calcium-independent_phospholipase_A2-gamma-like_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 3.585
Sequence
CDS
ATGTCTTTCGGAGGTCAGTGGCGCGTACTGCGACACTATTTGTCTCTAACAAATTTGAATGCAGAAGCAGAAAAATTAATTCAAAAAATTAAACCAGTTACACCAGAACAATGGGAAAGGTTCCTGACGAAACTAGAAAAAGCGTTGTCCGTCCAAGTAATAGAACAAAATAAAAGTCAAGAAGTAATAAATTTAAAGGACATTAGAGTCAAAGCGACCGCCATCGAGGAAACTCACAAAACCGAAAAGTCTGACGAGAGGCAGGCCGGTGGACTCGGCATCGGCTCAGAGGCAAAGGAAATAACATTCGAAAGCTTCCTTCAAGGGTTGAAGTTGAAAAAGGATTTGCATTTCGGCAAGCTATCGGAGCCAAGTTGGAAGTCTAACAAGCCAACTGTTACGAAGACATCGATTCATTCTCGGACTGCGTACGTCATATCCGCGATCGTAACTGCTGAAACTTCAGAATGCCTCATGAAAAGAACGGAACATTTCATAGATCACCTAACGCAGTATCCTGAAGCCAGAGATTACGCTATCAAGGAAGGTGCGGTCAGGGCGTTATTGCGCGTGGAAAACAAATTAAACGATGAAATTGAAATTGATAAACAAGTAAAAGGAGTCGTTAATGAGGCCCTCGCTCTGGTGGGCTACACGGGGCCCACTAAAGGACCGGGCCCTAACATCCTATCCCTGGACGGAGGTGGAATACGAGGTCTGATAGCCATAGAGATCCTAAAGCACCTCGAGAAGATAACGGGAAGGAAAGTGCACGAGCTCTTCGACTACATTATAGGCGTTAGTACGGGAGCCATCATCGCTGCTGTTATCGGCAGCGGAATCGGGTCACTCGAGACCGCGAGTCAGATGTACCGCACGCTGTCGAAGGAGATGTTCGGGAAGACCTCGGTCATCGGAGGCACCTCCAGGCTGGTCTGGACCCACTCGTACTACGACACCGAGGCCTGGGAGAAGATGCTGCAGGAGAATCTCAGGAACTGTACGCTCTCCGAGTGCAACCGGTACAACAGGCCGAAGATCGCGCTGGTGTCGTGCGTGGTGCGCCCGGGCGCGGGGCTGAGCCCGTACGTGTTCCGCTCGTACTCGTGCGGGTTCCGCGTGCGCGCCGCGCTGCCGGGCAGCGGCCGCCCCGCGCTGTGGCAGGCCGTGCGCGCCTCCGCCGCCGCGCCCACATACTTCCACGAGTTCACGCTGGACGGACTCATCCACCAGGATGGTGGTATTATGGTGAACAATCCTACCGGGGTAGGGATACACGAAGCCAGGCTACTATACGGAGCGGATGCCCTAAGGAAGGGCACCATCATATCTGTAGGAACAGGGAAGGCCCTCAACAAACACGCGGAGAACAGGGGTCTGAGTAAGGGCGCGCCAAAGGACCCTGCAGGGACCAGTTGGAAGGAGAAGTTCAACAAGATATTGGAGTCCGCCACTGATACCGAAGGAGTGCACCTGGTCGTGAGCGAGCTGTTGCCCCCGGGCTCTTACTACCGCTTCAACCCCCCGCTGATGCAGGAGTGCGCTATGGACGAGATCGACCCTGAGAAACTGGCCAGTCTGCTCACAGACACCAATGACTATATCAGACGGAACCAGCACAAGTTCGAGCGCGCGGCCGCGATGCTGACCCGGAAGCGCGGCCTCGTGCAGCGCGCGGCAGACTACGTGCGGCACAAGCGGCAGATCATGGGCCTAGTCCAGGGGCACTGA
Protein
MSFGGQWRVLRHYLSLTNLNAEAEKLIQKIKPVTPEQWERFLTKLEKALSVQVIEQNKSQEVINLKDIRVKATAIEETHKTEKSDERQAGGLGIGSEAKEITFESFLQGLKLKKDLHFGKLSEPSWKSNKPTVTKTSIHSRTAYVISAIVTAETSECLMKRTEHFIDHLTQYPEARDYAIKEGAVRALLRVENKLNDEIEIDKQVKGVVNEALALVGYTGPTKGPGPNILSLDGGGIRGLIAIEILKHLEKITGRKVHELFDYIIGVSTGAIIAAVIGSGIGSLETASQMYRTLSKEMFGKTSVIGGTSRLVWTHSYYDTEAWEKMLQENLRNCTLSECNRYNRPKIALVSCVVRPGAGLSPYVFRSYSCGFRVRAALPGSGRPALWQAVRASAAAPTYFHEFTLDGLIHQDGGIMVNNPTGVGIHEARLLYGADALRKGTIISVGTGKALNKHAENRGLSKGAPKDPAGTSWKEKFNKILESATDTEGVHLVVSELLPPGSYYRFNPPLMQECAMDEIDPEKLASLLTDTNDYIRRNQHKFERAAAMLTRKRGLVQRAADYVRHKRQIMGLVQGH
Summary
Uniprot
H9JWJ6
A0A0N1PF66
A0A2H1V4K8
M4M7W8
A0A212FDY7
M4M6X2
+ More
A0A3S2LRJ8 A0A2W1BAJ9 A0A2A4JKM3 A0A154PLG3 A0A0L7RI55 D6WQY5 E2B4C9 A0A2P8YJ87 A0A348G5X3 A0A0N0BDG8 K7JAC1 N6TT40 A0A026WC74 E9J756 A0A0T6BEM6 A0A1B6CNX1 E2APH6 A0A151J3Q2 A0A1B6CNR4 A0A195B5X2 A0A158P102 E0VMF2 A0A2J7PFQ8 A0A310SFS7 A0A2A3E488 A0A088AMQ5 A0A151IAF1 A0A067QMK2 D6WQY7 A0A195F469 A0A2J7PFR4 A0A1B6GSW1 A0A1W4WIB9 A0A1B6GS85 F4W993 A0A151X8X1 A0A0C9RKD9 A0A2J7PFR6 A0A0C9QGM7 A0A1Y1M601 A0A0P5RG32 A0A0P5EK72 A0A0N8AGZ4 A0A0P5SR04 A0A1V9X4B9 A0A0P5SXU7 A0A0P5SM16 A0A0N8BL93 A0A0P5MNE6 A0A0P5JRC4 A0A0P5XWU5 A0A0P5LJS4 A0A0P6HF19 A0A0P4W549 A0A0P6B7A3 T1JTZ1 T1ILI3 A0A0P5UCP5 A0A224Z2T8 A0A023GC31 A0A3M0KA69 L7MKP4 A0A293LIE6 A0A131YU09 A0A091N1P9 U3K6S6 A0A093JCA4 A0A091JZJ5 A0A0F7Z5B9 U3IKL5 A0A091GBX6 I3JPW7 A0A091PS00 A0A0A0A8S2 A0A094LF72 A0A131XI17 A0A2R5LHV6 A0A093NZX1 A0A091SW11 A0A087RGS7 A0A093QB28 A0A3P9NTH6 A0A099YSD0 A0A2D4IU80 A0A3R7PFI2 A0A2I0UJN3 R7U8N4 G1K846 A0A091P3X0 V9KEW4
A0A3S2LRJ8 A0A2W1BAJ9 A0A2A4JKM3 A0A154PLG3 A0A0L7RI55 D6WQY5 E2B4C9 A0A2P8YJ87 A0A348G5X3 A0A0N0BDG8 K7JAC1 N6TT40 A0A026WC74 E9J756 A0A0T6BEM6 A0A1B6CNX1 E2APH6 A0A151J3Q2 A0A1B6CNR4 A0A195B5X2 A0A158P102 E0VMF2 A0A2J7PFQ8 A0A310SFS7 A0A2A3E488 A0A088AMQ5 A0A151IAF1 A0A067QMK2 D6WQY7 A0A195F469 A0A2J7PFR4 A0A1B6GSW1 A0A1W4WIB9 A0A1B6GS85 F4W993 A0A151X8X1 A0A0C9RKD9 A0A2J7PFR6 A0A0C9QGM7 A0A1Y1M601 A0A0P5RG32 A0A0P5EK72 A0A0N8AGZ4 A0A0P5SR04 A0A1V9X4B9 A0A0P5SXU7 A0A0P5SM16 A0A0N8BL93 A0A0P5MNE6 A0A0P5JRC4 A0A0P5XWU5 A0A0P5LJS4 A0A0P6HF19 A0A0P4W549 A0A0P6B7A3 T1JTZ1 T1ILI3 A0A0P5UCP5 A0A224Z2T8 A0A023GC31 A0A3M0KA69 L7MKP4 A0A293LIE6 A0A131YU09 A0A091N1P9 U3K6S6 A0A093JCA4 A0A091JZJ5 A0A0F7Z5B9 U3IKL5 A0A091GBX6 I3JPW7 A0A091PS00 A0A0A0A8S2 A0A094LF72 A0A131XI17 A0A2R5LHV6 A0A093NZX1 A0A091SW11 A0A087RGS7 A0A093QB28 A0A3P9NTH6 A0A099YSD0 A0A2D4IU80 A0A3R7PFI2 A0A2I0UJN3 R7U8N4 G1K846 A0A091P3X0 V9KEW4
Pubmed
EMBL
BABH01018556
BABH01018557
KQ459166
KPJ03543.1
ODYU01000645
SOQ35768.1
+ More
JX846988 AGG55005.1 AGBW02008980 OWR51985.1 JX847002 AGG55019.1 RSAL01000292 RVE42891.1 KZ150272 PZC71671.1 NWSH01001099 PCG72607.1 KQ434960 KZC12673.1 KQ414590 KOC70411.1 KQ971351 EFA06489.1 GL445549 EFN89477.1 PYGN01000555 PSN44318.1 FX985510 BBF97846.1 KQ435876 KOX70148.1 AAZX01012324 APGK01055492 APGK01055493 KB741266 KB632194 ENN71511.1 ERL89934.1 KK107293 QOIP01000005 EZA53271.1 RLU22830.1 GL768421 EFZ11375.1 LJIG01001187 KRT85753.1 GEDC01022091 JAS15207.1 GL441542 EFN64675.1 KQ980275 KYN17033.1 GEDC01022343 JAS14955.1 KQ976598 KYM79589.1 ADTU01001034 DS235307 EEB14558.1 NEVH01025650 PNF15170.1 KQ760478 OAD60245.1 KZ288382 PBC26510.1 KQ978223 KYM96261.1 KK853244 KDR09436.1 EFA06525.2 KQ981855 KYN34879.1 PNF15172.1 GECZ01004336 GECZ01001990 JAS65433.1 JAS67779.1 GECZ01004479 JAS65290.1 GL888002 EGI69275.1 KQ982402 KYQ56836.1 GBYB01013732 JAG83499.1 PNF15171.1 GBYB01013733 JAG83500.1 GEZM01039817 JAV81163.1 GDIQ01120574 JAL31152.1 GDIP01147091 JAJ76311.1 GDIP01148385 JAJ75017.1 GDIP01136588 JAL67126.1 MNPL01024579 OQR68470.1 GDIP01136589 JAL67125.1 GDIP01138081 JAL65633.1 GDIQ01166715 JAK85010.1 GDIQ01165174 JAK86551.1 GDIQ01195104 JAK56621.1 GDIP01066099 LRGB01000389 JAM37616.1 KZS19389.1 GDIQ01168616 JAK83109.1 GDIQ01020370 JAN74367.1 GDRN01086916 GDRN01086915 GDRN01086914 JAI61108.1 GDIP01018662 JAM85053.1 CAEY01000486 JH430824 GDIP01116331 JAL87383.1 GFPF01009174 MAA20320.1 GBBM01003627 JAC31791.1 QRBI01000112 RMC10083.1 GACK01001300 JAA63734.1 GFWV01001755 MAA26485.1 GEDV01005848 JAP82709.1 KL374540 KFP82867.1 AGTO01020686 KK579548 KFW12670.1 KK536440 KFP29551.1 GBEW01000706 JAI09659.1 KL447924 KFO78796.1 AERX01010948 AERX01010949 KK664908 KFQ10662.1 KL870977 KGL89893.1 KL356665 KFZ62604.1 GEFH01001832 JAP66749.1 GGLE01004975 MBY09101.1 KL225237 KFW69671.1 KK487668 KFQ62493.1 KL226357 KFM12681.1 KL672442 KFW85791.1 KL885679 KGL73184.1 IACK01125971 LAA87736.1 QCYY01000271 ROT85468.1 KZ505720 PKU46262.1 AMQN01008807 KB304010 ELU02471.1 KK668043 KFQ02652.1 JW863859 AFO96376.1
JX846988 AGG55005.1 AGBW02008980 OWR51985.1 JX847002 AGG55019.1 RSAL01000292 RVE42891.1 KZ150272 PZC71671.1 NWSH01001099 PCG72607.1 KQ434960 KZC12673.1 KQ414590 KOC70411.1 KQ971351 EFA06489.1 GL445549 EFN89477.1 PYGN01000555 PSN44318.1 FX985510 BBF97846.1 KQ435876 KOX70148.1 AAZX01012324 APGK01055492 APGK01055493 KB741266 KB632194 ENN71511.1 ERL89934.1 KK107293 QOIP01000005 EZA53271.1 RLU22830.1 GL768421 EFZ11375.1 LJIG01001187 KRT85753.1 GEDC01022091 JAS15207.1 GL441542 EFN64675.1 KQ980275 KYN17033.1 GEDC01022343 JAS14955.1 KQ976598 KYM79589.1 ADTU01001034 DS235307 EEB14558.1 NEVH01025650 PNF15170.1 KQ760478 OAD60245.1 KZ288382 PBC26510.1 KQ978223 KYM96261.1 KK853244 KDR09436.1 EFA06525.2 KQ981855 KYN34879.1 PNF15172.1 GECZ01004336 GECZ01001990 JAS65433.1 JAS67779.1 GECZ01004479 JAS65290.1 GL888002 EGI69275.1 KQ982402 KYQ56836.1 GBYB01013732 JAG83499.1 PNF15171.1 GBYB01013733 JAG83500.1 GEZM01039817 JAV81163.1 GDIQ01120574 JAL31152.1 GDIP01147091 JAJ76311.1 GDIP01148385 JAJ75017.1 GDIP01136588 JAL67126.1 MNPL01024579 OQR68470.1 GDIP01136589 JAL67125.1 GDIP01138081 JAL65633.1 GDIQ01166715 JAK85010.1 GDIQ01165174 JAK86551.1 GDIQ01195104 JAK56621.1 GDIP01066099 LRGB01000389 JAM37616.1 KZS19389.1 GDIQ01168616 JAK83109.1 GDIQ01020370 JAN74367.1 GDRN01086916 GDRN01086915 GDRN01086914 JAI61108.1 GDIP01018662 JAM85053.1 CAEY01000486 JH430824 GDIP01116331 JAL87383.1 GFPF01009174 MAA20320.1 GBBM01003627 JAC31791.1 QRBI01000112 RMC10083.1 GACK01001300 JAA63734.1 GFWV01001755 MAA26485.1 GEDV01005848 JAP82709.1 KL374540 KFP82867.1 AGTO01020686 KK579548 KFW12670.1 KK536440 KFP29551.1 GBEW01000706 JAI09659.1 KL447924 KFO78796.1 AERX01010948 AERX01010949 KK664908 KFQ10662.1 KL870977 KGL89893.1 KL356665 KFZ62604.1 GEFH01001832 JAP66749.1 GGLE01004975 MBY09101.1 KL225237 KFW69671.1 KK487668 KFQ62493.1 KL226357 KFM12681.1 KL672442 KFW85791.1 KL885679 KGL73184.1 IACK01125971 LAA87736.1 QCYY01000271 ROT85468.1 KZ505720 PKU46262.1 AMQN01008807 KB304010 ELU02471.1 KK668043 KFQ02652.1 JW863859 AFO96376.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000283053
UP000218220
UP000076502
+ More
UP000053825 UP000007266 UP000008237 UP000245037 UP000053105 UP000002358 UP000019118 UP000030742 UP000053097 UP000279307 UP000000311 UP000078492 UP000078540 UP000005205 UP000009046 UP000235965 UP000242457 UP000005203 UP000078542 UP000027135 UP000078541 UP000192223 UP000007755 UP000075809 UP000192247 UP000076858 UP000015104 UP000269221 UP000016665 UP000053760 UP000005207 UP000053858 UP000054081 UP000053286 UP000053258 UP000242638 UP000053641 UP000283509 UP000014760 UP000001646
UP000053825 UP000007266 UP000008237 UP000245037 UP000053105 UP000002358 UP000019118 UP000030742 UP000053097 UP000279307 UP000000311 UP000078492 UP000078540 UP000005205 UP000009046 UP000235965 UP000242457 UP000005203 UP000078542 UP000027135 UP000078541 UP000192223 UP000007755 UP000075809 UP000192247 UP000076858 UP000015104 UP000269221 UP000016665 UP000053760 UP000005207 UP000053858 UP000054081 UP000053286 UP000053258 UP000242638 UP000053641 UP000283509 UP000014760 UP000001646
PRIDE
SUPFAM
SSF52151
SSF52151
ProteinModelPortal
H9JWJ6
A0A0N1PF66
A0A2H1V4K8
M4M7W8
A0A212FDY7
M4M6X2
+ More
A0A3S2LRJ8 A0A2W1BAJ9 A0A2A4JKM3 A0A154PLG3 A0A0L7RI55 D6WQY5 E2B4C9 A0A2P8YJ87 A0A348G5X3 A0A0N0BDG8 K7JAC1 N6TT40 A0A026WC74 E9J756 A0A0T6BEM6 A0A1B6CNX1 E2APH6 A0A151J3Q2 A0A1B6CNR4 A0A195B5X2 A0A158P102 E0VMF2 A0A2J7PFQ8 A0A310SFS7 A0A2A3E488 A0A088AMQ5 A0A151IAF1 A0A067QMK2 D6WQY7 A0A195F469 A0A2J7PFR4 A0A1B6GSW1 A0A1W4WIB9 A0A1B6GS85 F4W993 A0A151X8X1 A0A0C9RKD9 A0A2J7PFR6 A0A0C9QGM7 A0A1Y1M601 A0A0P5RG32 A0A0P5EK72 A0A0N8AGZ4 A0A0P5SR04 A0A1V9X4B9 A0A0P5SXU7 A0A0P5SM16 A0A0N8BL93 A0A0P5MNE6 A0A0P5JRC4 A0A0P5XWU5 A0A0P5LJS4 A0A0P6HF19 A0A0P4W549 A0A0P6B7A3 T1JTZ1 T1ILI3 A0A0P5UCP5 A0A224Z2T8 A0A023GC31 A0A3M0KA69 L7MKP4 A0A293LIE6 A0A131YU09 A0A091N1P9 U3K6S6 A0A093JCA4 A0A091JZJ5 A0A0F7Z5B9 U3IKL5 A0A091GBX6 I3JPW7 A0A091PS00 A0A0A0A8S2 A0A094LF72 A0A131XI17 A0A2R5LHV6 A0A093NZX1 A0A091SW11 A0A087RGS7 A0A093QB28 A0A3P9NTH6 A0A099YSD0 A0A2D4IU80 A0A3R7PFI2 A0A2I0UJN3 R7U8N4 G1K846 A0A091P3X0 V9KEW4
A0A3S2LRJ8 A0A2W1BAJ9 A0A2A4JKM3 A0A154PLG3 A0A0L7RI55 D6WQY5 E2B4C9 A0A2P8YJ87 A0A348G5X3 A0A0N0BDG8 K7JAC1 N6TT40 A0A026WC74 E9J756 A0A0T6BEM6 A0A1B6CNX1 E2APH6 A0A151J3Q2 A0A1B6CNR4 A0A195B5X2 A0A158P102 E0VMF2 A0A2J7PFQ8 A0A310SFS7 A0A2A3E488 A0A088AMQ5 A0A151IAF1 A0A067QMK2 D6WQY7 A0A195F469 A0A2J7PFR4 A0A1B6GSW1 A0A1W4WIB9 A0A1B6GS85 F4W993 A0A151X8X1 A0A0C9RKD9 A0A2J7PFR6 A0A0C9QGM7 A0A1Y1M601 A0A0P5RG32 A0A0P5EK72 A0A0N8AGZ4 A0A0P5SR04 A0A1V9X4B9 A0A0P5SXU7 A0A0P5SM16 A0A0N8BL93 A0A0P5MNE6 A0A0P5JRC4 A0A0P5XWU5 A0A0P5LJS4 A0A0P6HF19 A0A0P4W549 A0A0P6B7A3 T1JTZ1 T1ILI3 A0A0P5UCP5 A0A224Z2T8 A0A023GC31 A0A3M0KA69 L7MKP4 A0A293LIE6 A0A131YU09 A0A091N1P9 U3K6S6 A0A093JCA4 A0A091JZJ5 A0A0F7Z5B9 U3IKL5 A0A091GBX6 I3JPW7 A0A091PS00 A0A0A0A8S2 A0A094LF72 A0A131XI17 A0A2R5LHV6 A0A093NZX1 A0A091SW11 A0A087RGS7 A0A093QB28 A0A3P9NTH6 A0A099YSD0 A0A2D4IU80 A0A3R7PFI2 A0A2I0UJN3 R7U8N4 G1K846 A0A091P3X0 V9KEW4
PDB
6AUN
E-value=1.63803e-15,
Score=203
Ontologies
GO
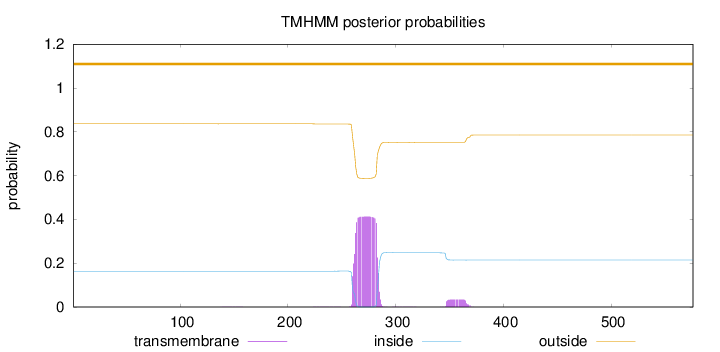
Topology
Length:
576
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.90806999999996
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.16341
outside
1 - 576
Population Genetic Test Statistics
Pi
222.68976
Theta
163.701862
Tajima's D
1.052799
CLR
0.568521
CSRT
0.673416329183541
Interpretation
Uncertain
